www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
We used D. melanogaster pigmentation as a focal trait to explore parallelism in phenotypic and genomic responses to environmental change - read more at the link below 👇
doi.org/10.1093/evle...
Now in @evolletters.bsky.social by @skylerberardi.bsky.social, @paulrschmidt.bsky.social et al.
📷: Dr. Rush Dhillon
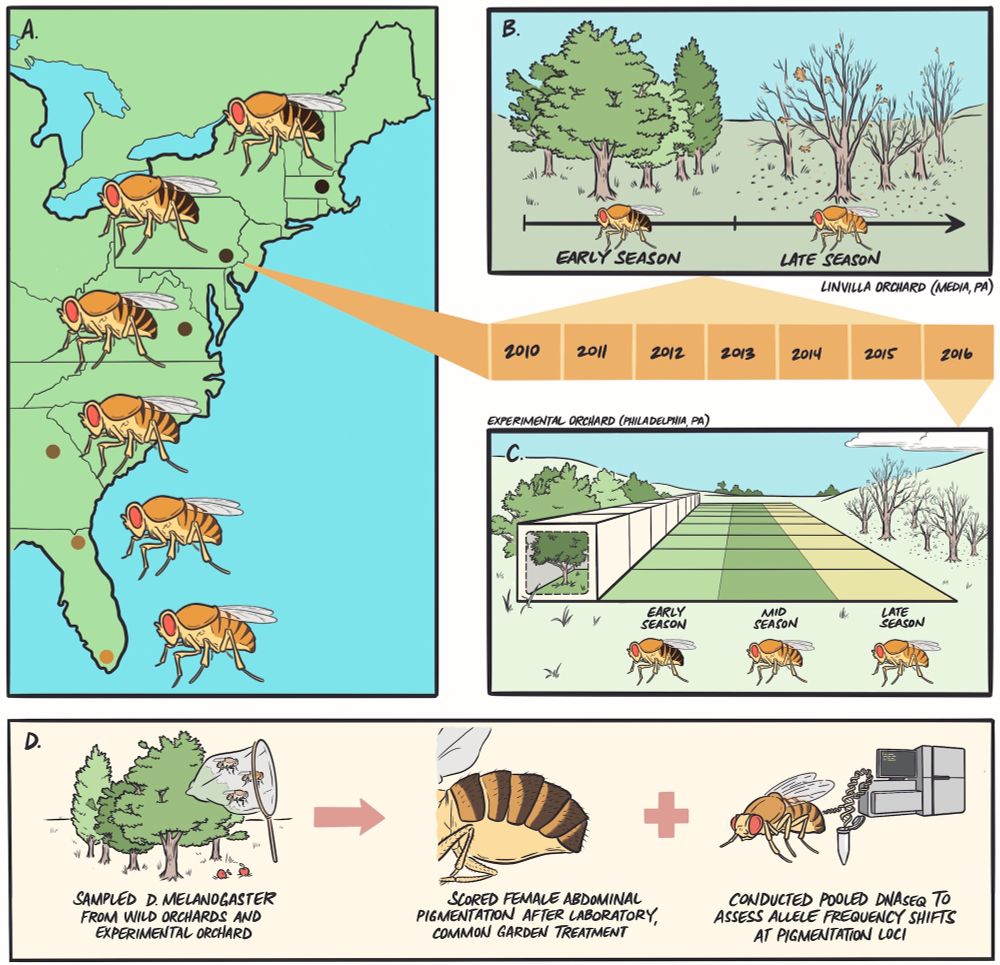
We used D. melanogaster pigmentation as a focal trait to explore parallelism in phenotypic and genomic responses to environmental change - read more at the link below 👇




As you know @flybase.bsky.social has been under financial pressure due to #NIH cuts for several years.
With the threats Harvard is currently facing, supporting FlyBase is even more essential. If you have the means, make a tax-deductible donation.
As you know @flybase.bsky.social has been under financial pressure due to #NIH cuts for several years.
With the threats Harvard is currently facing, supporting FlyBase is even more essential. If you have the means, make a tax-deductible donation.


Excited to share FlyCADD: impact prediction tool scoring the functional impact of any single nucleotide variant across the entire Drosophila melanogaster genome, to distinguish causal from neutral SNPs
www.biorxiv.org/content/10.1...

Excited to share FlyCADD: impact prediction tool scoring the functional impact of any single nucleotide variant across the entire Drosophila melanogaster genome, to distinguish causal from neutral SNPs
www.biorxiv.org/content/10.1...
Thrilled to share our work showing how beneficial dominance reversal helps fruit flies maintain a resistance polymorphism as selection varies in their environment! A thread 🧵 1/n
www.biorxiv.org/content/10.1...

Thrilled to share our work showing how beneficial dominance reversal helps fruit flies maintain a resistance polymorphism as selection varies in their environment! A thread 🧵 1/n
www.biorxiv.org/content/10.1...

@petrovadmitri.bsky.social, and others, we use low-input (down to 30ng), single-insect Nanopore sequencing to help do 184 new genomes and address the near absence of genomic data for drosophilid species that cannot be cultured in the lab.

@petrovadmitri.bsky.social, and others, we use low-input (down to 30ng), single-insect Nanopore sequencing to help do 184 new genomes and address the near absence of genomic data for drosophilid species that cannot be cultured in the lab.

