Interested in understanding the relationship between 3D genome structure and function
www.nature.com/articles/s41...

www.nature.com/articles/s41...
Simulations from @irate-physicist.bsky.social point to compaction.
Mitotic chr 2-3x more compacted --> lower configurational entropy --> lower entropic penalty for forming microcompartments --> microcompartment formation favored in mitosis!

Simulations from @irate-physicist.bsky.social point to compaction.
Mitotic chr 2-3x more compacted --> lower configurational entropy --> lower entropic penalty for forming microcompartments --> microcompartment formation favored in mitosis!
Prior work from @chrishsiung.bsky.social Gerd Blobel discovered 'mitotic transcriptional spiking', where ~half of genes transcriptionally spike in mitosis.
We now show that microcompartment dot number and strength predicts spiking

Prior work from @chrishsiung.bsky.social Gerd Blobel discovered 'mitotic transcriptional spiking', where ~half of genes transcriptionally spike in mitosis.
We now show that microcompartment dot number and strength predicts spiking
www.nature.com/articles/s41...
www.nature.com/articles/s41...
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...


www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
I know of earlier single-locus papers (1982).
www.sciencedirect.com/science/arti...

I know of earlier single-locus papers (1982).
www.sciencedirect.com/science/arti...
www.science.org/doi/10.1126/...


www.science.org/doi/10.1126/...
STAG3-cohesin has a much shorter residence time which leads to altered 3D genome organization and STAG3-cohesin is important for male germ cell differentiation.
www.nature.com/articles/s41...

STAG3-cohesin has a much shorter residence time which leads to altered 3D genome organization and STAG3-cohesin is important for male germ cell differentiation.
www.nature.com/articles/s41...
www.nature.com/articles/s41...
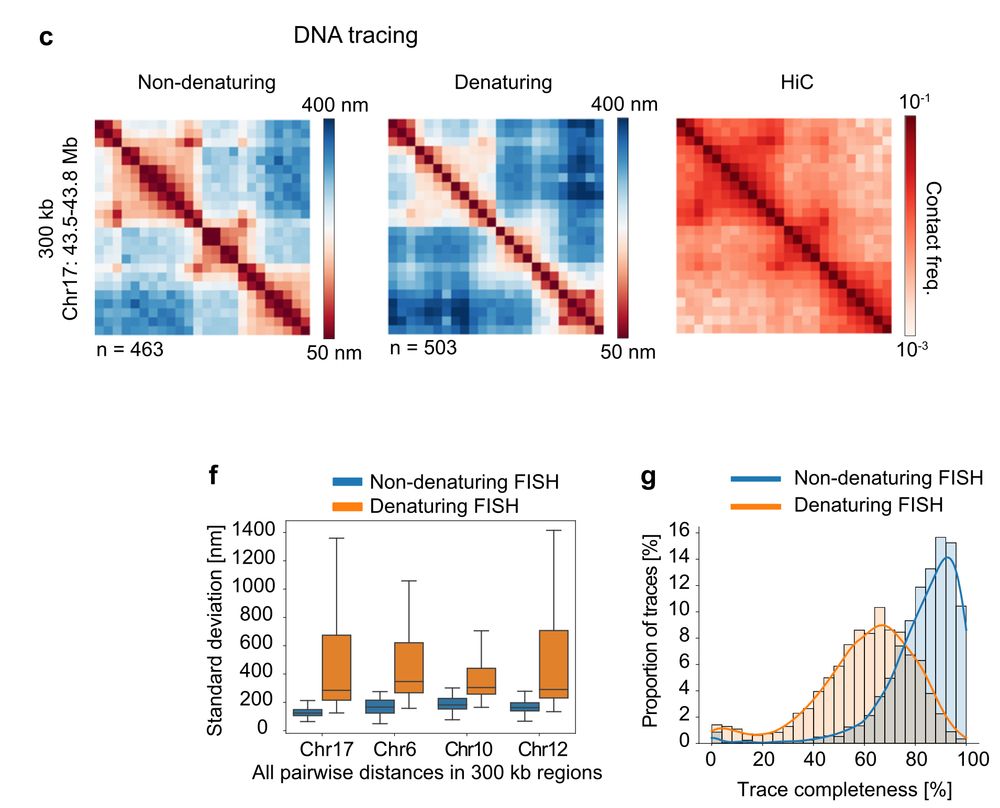
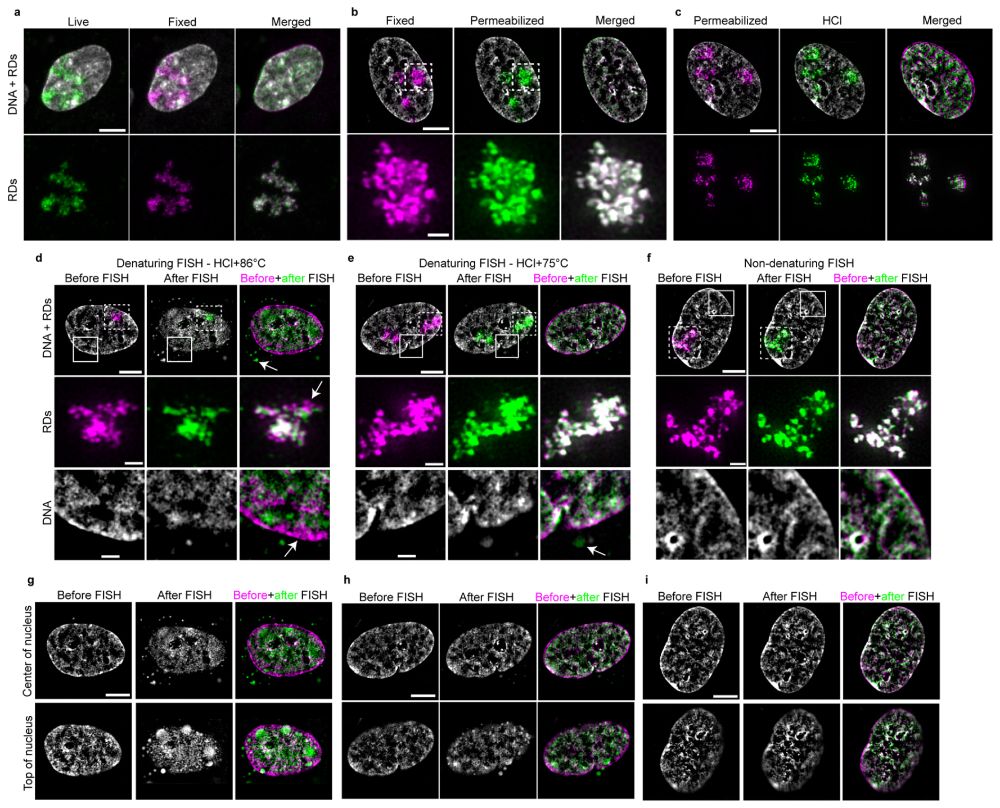
www.nature.com/articles/s41...


Thanks to Abberior - MINFLUX is awesome!
We would love critical feedback.
Full preprint here www.biorxiv.org/content/10.1...


Thanks to Abberior - MINFLUX is awesome!
We would love critical feedback.
Full preprint here www.biorxiv.org/content/10.1...
Strong subdiffusion makes frequent contact inevitable for E-P separations of ~200 nm.
Frequent local CRE contacts also predict microcompartments

Strong subdiffusion makes frequent contact inevitable for E-P separations of ~200 nm.
Frequent local CRE contacts also predict microcompartments
- Nucleus is crowded (~400 genes + ~8,000 per um^3)
- Finding nearby (within few hundred kb) enhancers is very efficient
- Very distal and generally non-cognate enhancers (>Mb) are avoided unless facilitated by other mechanisms (e.g. loop extrusion)

- Nucleus is crowded (~400 genes + ~8,000 per um^3)
- Finding nearby (within few hundred kb) enhancers is very efficient
- Very distal and generally non-cognate enhancers (>Mb) are avoided unless facilitated by other mechanisms (e.g. loop extrusion)
Rather, it makes finding nearby things extremely efficient and finding very distal things nearly impossible.
median first passage time estimates:
~4 ms to find 100 nm distal target
>25 hours to find 1300 nm distal target

Rather, it makes finding nearby things extremely efficient and finding very distal things nearly impossible.
median first passage time estimates:
~4 ms to find 100 nm distal target
>25 hours to find 1300 nm distal target
Only loop extrusion affects exponent.

Only loop extrusion affects exponent.
alpha~0.3 in U2OS cells across full 6+ OOM
mESC chromatin dynamics does not follow power law, invalidating mESC descriptions using single exponent 🤔
Both mESC+U2OS show strong subdiffusion

alpha~0.3 in U2OS cells across full 6+ OOM
mESC chromatin dynamics does not follow power law, invalidating mESC descriptions using single exponent 🤔
Both mESC+U2OS show strong subdiffusion
MINFLUX @stefanhelllabs.bsky.social is transformative and allows tracking at microsecond timescale, but procedure introduces bias for recurrent motion --> by carefully simulating MINFLUX to optimize L we can avoid this bias.

MINFLUX @stefanhelllabs.bsky.social is transformative and allows tracking at microsecond timescale, but procedure introduces bias for recurrent motion --> by carefully simulating MINFLUX to optimize L we can avoid this bias.
- 3 techniques: MINFLUX, SPT, SRLCI
- 2 labels: Histone H2B (global, pioneered by @kazu-maeshima.bsky.social ) and Fbn2 locus (local, www.science.org/doi/full/10.... )
- 2 cell lines: human U2OS, mESCs
- 4 perturbations: Loop extrusion, Transcription, Histone Acetylation, Supercoiling
- 3 techniques: MINFLUX, SPT, SRLCI
- 2 labels: Histone H2B (global, pioneered by @kazu-maeshima.bsky.social ) and Fbn2 locus (local, www.science.org/doi/full/10.... )
- 2 cell lines: human U2OS, mESCs
- 4 perturbations: Loop extrusion, Transcription, Histone Acetylation, Supercoiling
By integrating MINFLUX, we span 0.0002 to 7200 sec reaching range of 36,000,000 to span 7 OOM in time to map chromatin dynamics in live cells

By integrating MINFLUX, we span 0.0002 to 7200 sec reaching range of 36,000,000 to span 7 OOM in time to map chromatin dynamics in live cells
MSD~time^alpha:
If a~0.4 (fractal globule), need 5 OOM in time
If a~0.3, need 7 OOM in time
7 OOM in time means a movie with >10,000,000 frames 😱

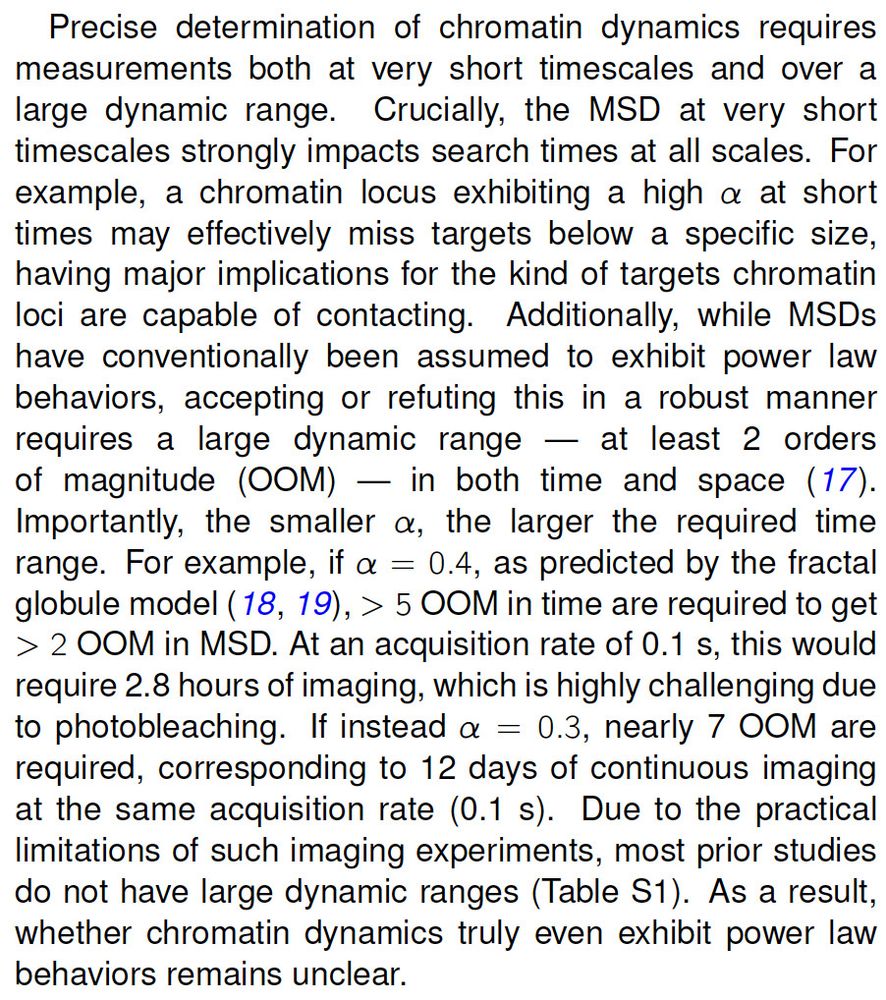
MSD~time^alpha:
If a~0.4 (fractal globule), need 5 OOM in time
If a~0.3, need 7 OOM in time
7 OOM in time means a movie with >10,000,000 frames 😱
Increase E-P separation from 10 to 500 kb:
if a~0.8, search time increases 300-fold
if a~0.2, search time increases 10-billion fold
Cannot understand E-P dynamics without alpha

Increase E-P separation from 10 to 500 kb:
if a~0.8, search time increases 300-fold
if a~0.2, search time increases 10-billion fold
Cannot understand E-P dynamics without alpha
Mean Squared Displacement (MSD) should follow power law: MSD ~ time^alpha
fBm simulation shows this: a~0.25 oversamples local environment, whereas Brownian motion a=1 explores more globally.
Mean Squared Displacement (MSD) should follow power law: MSD ~ time^alpha
fBm simulation shows this: a~0.25 oversamples local environment, whereas Brownian motion a=1 explores more globally.
Q: how does chromatin move?
Using MINFLUX, SPT & SRLCI, we track chromatin dynamics across 7 orders of magnitude in time to provide answers www.biorxiv.org/content/10.1...

Q: how does chromatin move?
Using MINFLUX, SPT & SRLCI, we track chromatin dynamics across 7 orders of magnitude in time to provide answers www.biorxiv.org/content/10.1...

