
AlphaFold Unofficial
@alphafold.bsky.social
Unofficial account exploring the intersection of biology, molecules, science, AI and protein folding with AlphaFold.
Pinned
By far the best scientific community on the planet. Feels great leaving Mordor for Bluer Skies! #science
Reposted by AlphaFold Unofficial
AlphaFold is pretty legit! I would say amino acid/nucleotide sequences are some of the best use cases. But in general the AI hype is dumb. If you don't tell me*how* you use AI in the pitch I'm assuming you shoehorned it in unnecessarily.
November 8, 2025 at 1:34 AM
AlphaFold is pretty legit! I would say amino acid/nucleotide sequences are some of the best use cases. But in general the AI hype is dumb. If you don't tell me*how* you use AI in the pitch I'm assuming you shoehorned it in unnecessarily.
We dream of simulated proteins, trust worthy AI and technology that heals and gives more than it takes.
November 7, 2025 at 9:40 PM
We dream of simulated proteins, trust worthy AI and technology that heals and gives more than it takes.
Reposted by AlphaFold Unofficial
If Linux (for OS) is any model, OpenFold3 wants to become the standard for structural biology in academia & industry alike. Ambitious—but necessary. #OpenSource https://www.maffulli.net/2025/10/30/openfold3-open-source-replacement-alphafold/ #ai

OpenFold3 is an Open Source foundation model competing with the proprietary AlphaFold - ][ Stefano Maffulli
Reposted OpenFold Consortium Releases Preview of OpenFold3: An Open-Source Foundation Model for Structure Prediction of Proteins, Nucleic Acids, and Drugs (businesswire.com) It was bound to …
www.maffulli.net
November 3, 2025 at 8:47 PM
If Linux (for OS) is any model, OpenFold3 wants to become the standard for structural biology in academia & industry alike. Ambitious—but necessary. #OpenSource https://www.maffulli.net/2025/10/30/openfold3-open-source-replacement-alphafold/ #ai
Reposted by AlphaFold Unofficial
Brian Weitzner, co-founder of OpenFold, discusses the collaborative effort to develop software and AI tools to predict the 3D structure of proteins, how its open licensing model ensures broad accessibility, and how it stacks up against AlphaFold.
thebioreport.podbean.com/e/an-ai-coll...
thebioreport.podbean.com/e/an-ai-coll...

November 5, 2025 at 3:24 PM
Brian Weitzner, co-founder of OpenFold, discusses the collaborative effort to develop software and AI tools to predict the 3D structure of proteins, how its open licensing model ensures broad accessibility, and how it stacks up against AlphaFold.
thebioreport.podbean.com/e/an-ai-coll...
thebioreport.podbean.com/e/an-ai-coll...
Reposted by AlphaFold Unofficial
OpenFold3: Open-source protein structure AI aims to match AlphaFold! Has anyone tried it yet? 🙌
www.nature.com/articles/d41...
github.com/aqlaboratory...
#microsky #bioinformatics
🧪 🧬 🖥️ ✨
www.nature.com/articles/d41...
github.com/aqlaboratory...
#microsky #bioinformatics
🧪 🧬 🖥️ ✨

Open-source protein structure AI aims to match AlphaFold
The developers of OpenFold3 have released an early version of the tool, which they hope will one day perform on par with DeepMind’s protein-structure model.
www.nature.com
November 6, 2025 at 5:25 AM
OpenFold3: Open-source protein structure AI aims to match AlphaFold! Has anyone tried it yet? 🙌
www.nature.com/articles/d41...
github.com/aqlaboratory...
#microsky #bioinformatics
🧪 🧬 🖥️ ✨
www.nature.com/articles/d41...
github.com/aqlaboratory...
#microsky #bioinformatics
🧪 🧬 🖥️ ✨
Reposted by AlphaFold Unofficial
Once you use AlphaFold to invent a new drug, it still has to be brought to market using pharma company infrastructure, not a server farm.
November 6, 2025 at 1:58 PM
Once you use AlphaFold to invent a new drug, it still has to be brought to market using pharma company infrastructure, not a server farm.
Human betterment will advance when biotechnological systems begin to read the body’s informational structure as an active language. Proteins, signals, and genetic loops can become co-authors in regenerative design through high-fidelity molecular modeling.
November 3, 2025 at 4:28 PM
Human betterment will advance when biotechnological systems begin to read the body’s informational structure as an active language. Proteins, signals, and genetic loops can become co-authors in regenerative design through high-fidelity molecular modeling.
When AI begins to simulate enzymatic catalysis at atomic resolution, biology transitions from observation to design. Protein structures cease to be static outputs and become programmable frameworks for therapeutic and environmental repair.
November 2, 2025 at 5:38 PM
When AI begins to simulate enzymatic catalysis at atomic resolution, biology transitions from observation to design. Protein structures cease to be static outputs and become programmable frameworks for therapeutic and environmental repair.
Reposted by AlphaFold Unofficial
This is going to be one of the biggest, most important features of AlphaFold @alphafold.bsky.social if further improved! Glycobiochemistry needs this bestfriend! #glycotime
academic.oup.com/glycob/artic...
academic.oup.com/glycob/artic...

Modeling glycans with AlphaFold 3: capabilities, caveats, and limitations
Abstract. Glycans are complex carbohydrates that exhibit extraordinary structural complexity and stereochemical diversity while playing essential roles in
academic.oup.com
October 31, 2025 at 8:51 PM
This is going to be one of the biggest, most important features of AlphaFold @alphafold.bsky.social if further improved! Glycobiochemistry needs this bestfriend! #glycotime
academic.oup.com/glycob/artic...
academic.oup.com/glycob/artic...
Reposted by AlphaFold Unofficial
Open-source protein structure AI aims to match AlphaFold
The system, called OpenFold3. OpenFold3 uses proteins’ amino acid sequences to map their 3D structures and model how they interact with other molecules, such as drugs or DNA.
The tool still doesn’t have the same functionality as […]
The system, called OpenFold3. OpenFold3 uses proteins’ amino acid sequences to map their 3D structures and model how they interact with other molecules, such as drugs or DNA.
The tool still doesn’t have the same functionality as […]
Original post on mstdn.science
mstdn.science
October 29, 2025 at 6:17 AM
Open-source protein structure AI aims to match AlphaFold
The system, called OpenFold3. OpenFold3 uses proteins’ amino acid sequences to map their 3D structures and model how they interact with other molecules, such as drugs or DNA.
The tool still doesn’t have the same functionality as […]
The system, called OpenFold3. OpenFold3 uses proteins’ amino acid sequences to map their 3D structures and model how they interact with other molecules, such as drugs or DNA.
The tool still doesn’t have the same functionality as […]
Reposted by AlphaFold Unofficial
Open-source protein structure AI aims to match AlphaFold www.nature.com/articles/d41...
Why open source is important for AI sovereignty and more, from a discussion at the @mediamuseum.bsky.social blog.scienceandmediamuseum.org.uk/how-britain-...
Why open source is important for AI sovereignty and more, from a discussion at the @mediamuseum.bsky.social blog.scienceandmediamuseum.org.uk/how-britain-...

Open-source protein structure AI aims to match AlphaFold
The developers of OpenFold3 have released an early version of the tool, which they hope will one day perform on par with DeepMind’s protein-structure model.
www.nature.com
October 29, 2025 at 10:56 AM
Open-source protein structure AI aims to match AlphaFold www.nature.com/articles/d41...
Why open source is important for AI sovereignty and more, from a discussion at the @mediamuseum.bsky.social blog.scienceandmediamuseum.org.uk/how-britain-...
Why open source is important for AI sovereignty and more, from a discussion at the @mediamuseum.bsky.social blog.scienceandmediamuseum.org.uk/how-britain-...
Reposted by AlphaFold Unofficial
L'IA open source de structure protéique vise à égaler AlphaFold

Open-source protein structure AI aims to match AlphaFold
The developers of OpenFold3 have released an early version of the tool, which they hope will one day perform on par with DeepMind’s protein-structure model.
www.nature.com
October 29, 2025 at 7:18 PM
L'IA open source de structure protéique vise à égaler AlphaFold
Reposted by AlphaFold Unofficial
This kind of consortium-led effort — academic + industry + nonprofit — shows how open collaboration can deliver foundational infrastructure for biology and AI, not just apps. 🤝 #OpenSource https://www.maffulli.net/2025/10/30/openfold3-open-source-replacement-alphafold/

OpenFold3 is an Open Source foundation model competing with the proprietary AlphaFold - ][ Stefano Maffulli
Reposted It was bound to happen: A challenge to the proprietary AlphaFold3 with full building code, training datasets, training checkpoints, developed by a non-profit consortium …
www.maffulli.net
October 30, 2025 at 11:53 AM
This kind of consortium-led effort — academic + industry + nonprofit — shows how open collaboration can deliver foundational infrastructure for biology and AI, not just apps. 🤝 #OpenSource https://www.maffulli.net/2025/10/30/openfold3-open-source-replacement-alphafold/
Reposted by AlphaFold Unofficial
AlphaFold DB v6 is here! 🚀
Heads up on key updates:
🔹 Website now only serves v6
🔹 New URL format!!
🔹 Dual API active for 9 months (retirement: 25-06-2026)
Use the AlphaFold API for reliable access. Time to update those bookmarks & scripts!
Heads up on key updates:
🔹 Website now only serves v6
🔹 New URL format!!
🔹 Dual API active for 9 months (retirement: 25-06-2026)
Use the AlphaFold API for reliable access. Time to update those bookmarks & scripts!
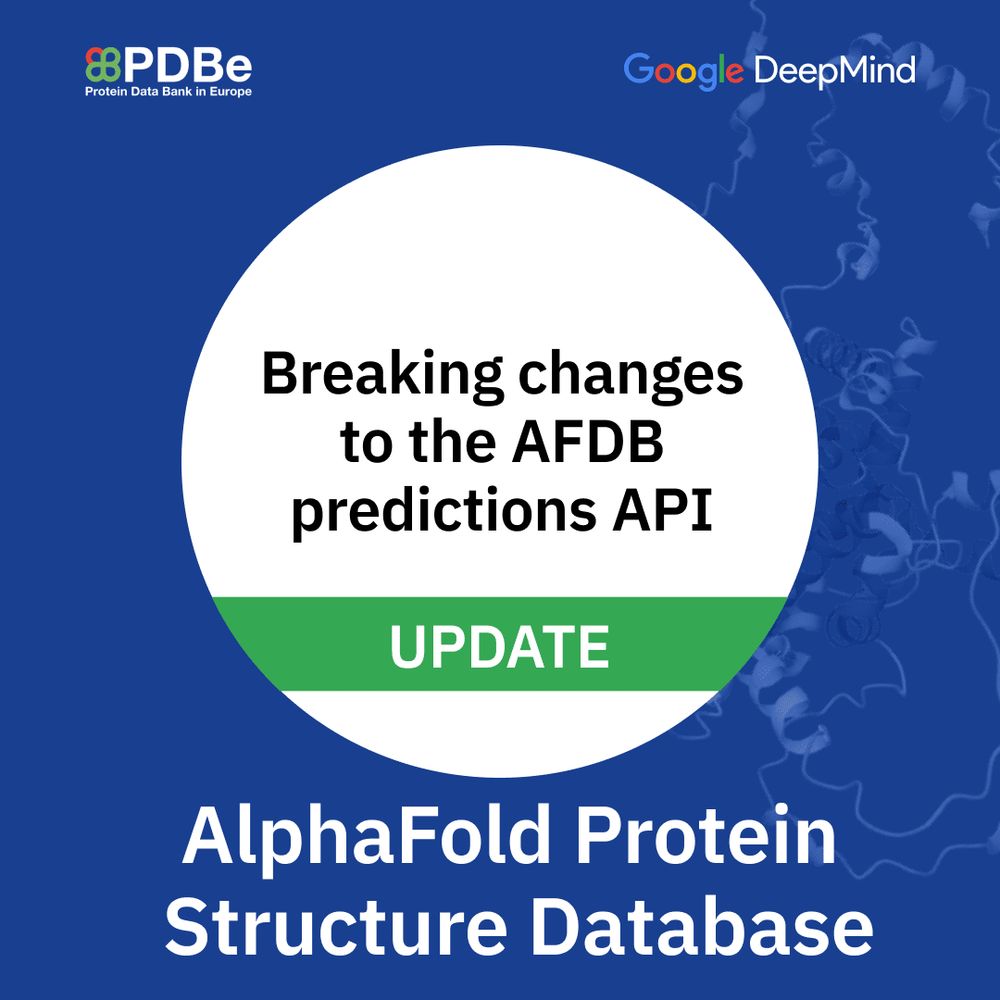
📣Important update for #AlphaFold API users! New API fields launched on October 7th, 2025. We’ll maintain BOTH fields until June 25, 2026! Check the docs! 🔗
For more information, read the news item: www.ebi.ac.uk/pdbe/news/br...
For more information, read the news item: www.ebi.ac.uk/pdbe/news/br...
October 22, 2025 at 5:39 PM
AlphaFold DB v6 is here! 🚀
Heads up on key updates:
🔹 Website now only serves v6
🔹 New URL format!!
🔹 Dual API active for 9 months (retirement: 25-06-2026)
Use the AlphaFold API for reliable access. Time to update those bookmarks & scripts!
Heads up on key updates:
🔹 Website now only serves v6
🔹 New URL format!!
🔹 Dual API active for 9 months (retirement: 25-06-2026)
Use the AlphaFold API for reliable access. Time to update those bookmarks & scripts!
Reposted by AlphaFold Unofficial
Any news of alphaproteo ? It's been over a year and since then, nothing. #alphafold
deepmind.google/discover/blo...
deepmind.google/discover/blo...

AlphaProteo generates novel proteins for biology and health research
New AI system designs proteins that successfully bind to target molecules, with potential for advancing drug design, disease understanding and more.
deepmind.google
October 24, 2025 at 9:51 AM
Any news of alphaproteo ? It's been over a year and since then, nothing. #alphafold
deepmind.google/discover/blo...
deepmind.google/discover/blo...
Reposted by AlphaFold Unofficial
This reminded me of what Dame Janet Thornton said about AlphaFold. It's a help to protein research, but not a substitute for protein researchers.
youtu.be/lxgaILSZEbU?...
youtu.be/lxgaILSZEbU?...

Ri on AI: Understanding AlphaFold – with Dame Janet Thornton
YouTube video by The Royal Institution
youtu.be
October 24, 2025 at 9:19 PM
This reminded me of what Dame Janet Thornton said about AlphaFold. It's a help to protein research, but not a substitute for protein researchers.
youtu.be/lxgaILSZEbU?...
youtu.be/lxgaILSZEbU?...
Reposted by AlphaFold Unofficial
I love AlphaFold—but please include PAE (Predicted Aligned Error) plots for every “interaction.” Pretty PDBs ≠ proof. If the PAE doesn’t show an interface, it ain’t one. Show the plots. Structural biology's reputation is on the line.
October 24, 2025 at 5:44 PM
I love AlphaFold—but please include PAE (Predicted Aligned Error) plots for every “interaction.” Pretty PDBs ≠ proof. If the PAE doesn’t show an interface, it ain’t one. Show the plots. Structural biology's reputation is on the line.
Reposted by AlphaFold Unofficial
Let's differentiate between GenAI and machine learning (ML).
Alphafold is ML. I'm personally familiar with several applications of ML that are transformational for biochemistry, drug development, and healthcare. ML is great at mining huge datasets for patterns.
I don't know anything about GenAI.
Alphafold is ML. I'm personally familiar with several applications of ML that are transformational for biochemistry, drug development, and healthcare. ML is great at mining huge datasets for patterns.
I don't know anything about GenAI.
October 17, 2025 at 2:48 PM
Let's differentiate between GenAI and machine learning (ML).
Alphafold is ML. I'm personally familiar with several applications of ML that are transformational for biochemistry, drug development, and healthcare. ML is great at mining huge datasets for patterns.
I don't know anything about GenAI.
Alphafold is ML. I'm personally familiar with several applications of ML that are transformational for biochemistry, drug development, and healthcare. ML is great at mining huge datasets for patterns.
I don't know anything about GenAI.
Reposted by AlphaFold Unofficial
AlphaFold heads to the clinic. DeepMind says three therapies for rare genetic diseases start trials in October 2025. With 200 million protein structures predicted by 2023, timelines could fall from 5 years to under 1, with cost savings. Big potential and big questions.
October 17, 2025 at 5:36 PM
AlphaFold heads to the clinic. DeepMind says three therapies for rare genetic diseases start trials in October 2025. With 200 million protein structures predicted by 2023, timelines could fall from 5 years to under 1, with cost savings. Big potential and big questions.
Reposted by AlphaFold Unofficial
This paper is incredible; IMO large-scale, unbiased data is our best chance to get beyond the relatively small set of sequences that AlphaFold loves
Our latest work seeks to answer a longstanding question: why is discovering new protein binders seemingly unpredictable – and can we better quantify and understand the de novo binder discover process? 1/12
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

Mapping the diverse topologies of protein-protein interaction fitness landscapes
De novo binder discovery is unpredictable and inefficient due to a lack of quantitative understanding of protein-protein interaction (PPI) sequence-function landscapes. Here, we use our PANCS-Binder t...
www.biorxiv.org
October 17, 2025 at 7:24 PM
This paper is incredible; IMO large-scale, unbiased data is our best chance to get beyond the relatively small set of sequences that AlphaFold loves
Reposted by AlphaFold Unofficial
Is the alphafold database performing really poorly, and Uniprot alphafold links broken for everyone, or just me?
I've tried clearing my cache, but any tips @ebi.embl.org?
I've tried clearing my cache, but any tips @ebi.embl.org?
October 18, 2025 at 3:49 AM
Is the alphafold database performing really poorly, and Uniprot alphafold links broken for everyone, or just me?
I've tried clearing my cache, but any tips @ebi.embl.org?
I've tried clearing my cache, but any tips @ebi.embl.org?
Reposted by AlphaFold Unofficial
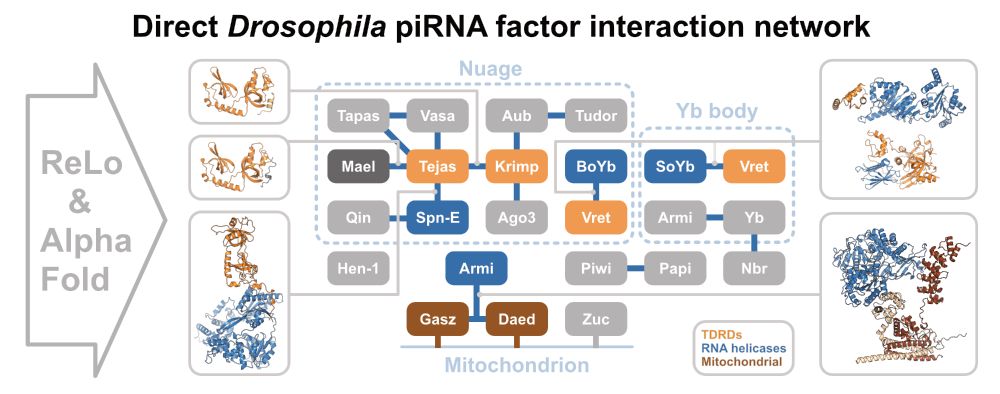
Excited to share our new @narjournal.bsky.social paper!
Using ReLo and AlphaFold, we described a network of interactions between Drosophila piRNA pathway factors suggesting new links how they coordinate to ensure transposon silencing and genome integrity.
academic.oup.com/nar/article/...
Using ReLo and AlphaFold, we described a network of interactions between Drosophila piRNA pathway factors suggesting new links how they coordinate to ensure transposon silencing and genome integrity.
academic.oup.com/nar/article/...
October 18, 2025 at 6:59 AM
Excited to share our new @narjournal.bsky.social paper!
Using ReLo and AlphaFold, we described a network of interactions between Drosophila piRNA pathway factors suggesting new links how they coordinate to ensure transposon silencing and genome integrity.
academic.oup.com/nar/article/...
Using ReLo and AlphaFold, we described a network of interactions between Drosophila piRNA pathway factors suggesting new links how they coordinate to ensure transposon silencing and genome integrity.
academic.oup.com/nar/article/...


