
onlinelibrary.wiley.com/doi/full/10....

onlinelibrary.wiley.com/doi/full/10....
bit.ly/plaid-proteins
🧵
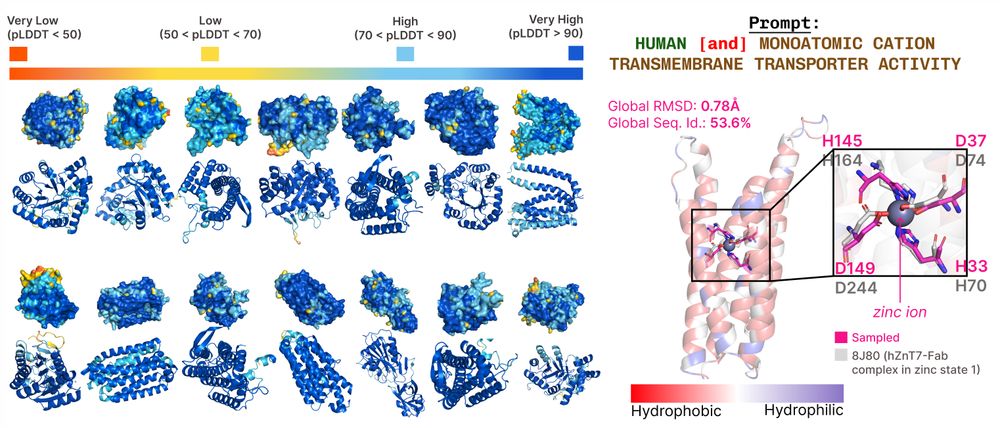
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
Check it out! The 300M and 600M models are open weights, 300M available for commercial usage.
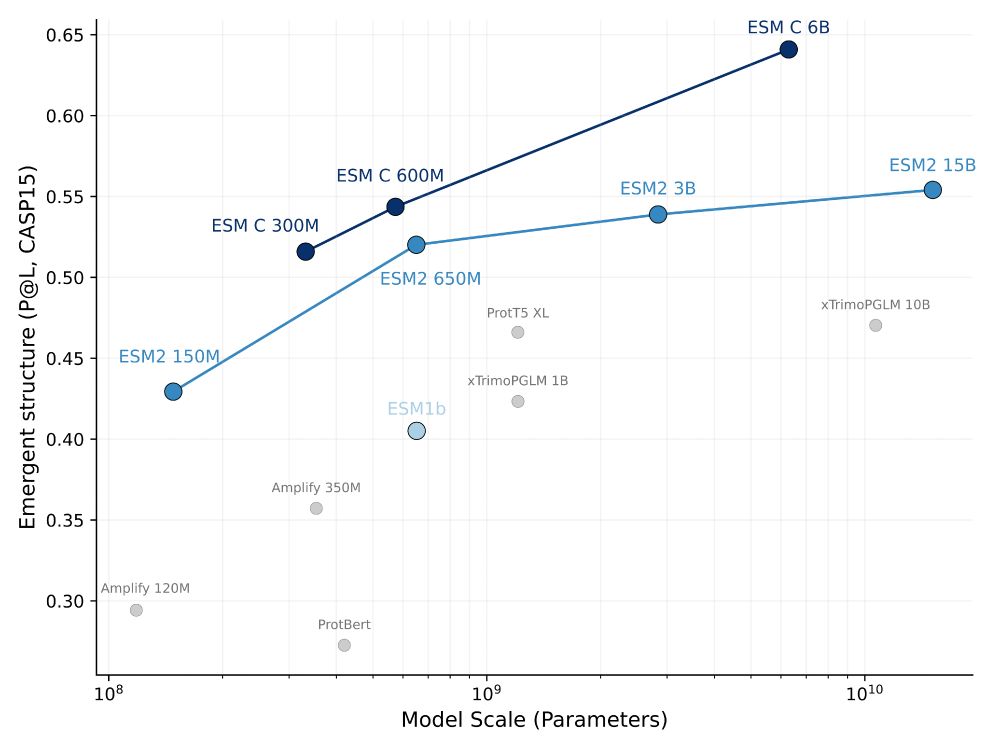
Check it out! The 300M and 600M models are open weights, 300M available for commercial usage.
www.businesswire.com/news/home/20...

www.businesswire.com/news/home/20...
-Physicist Fritz Houtermans
There's a lot of truth to this. log-log plots are often abused and can be very misleading
1/5
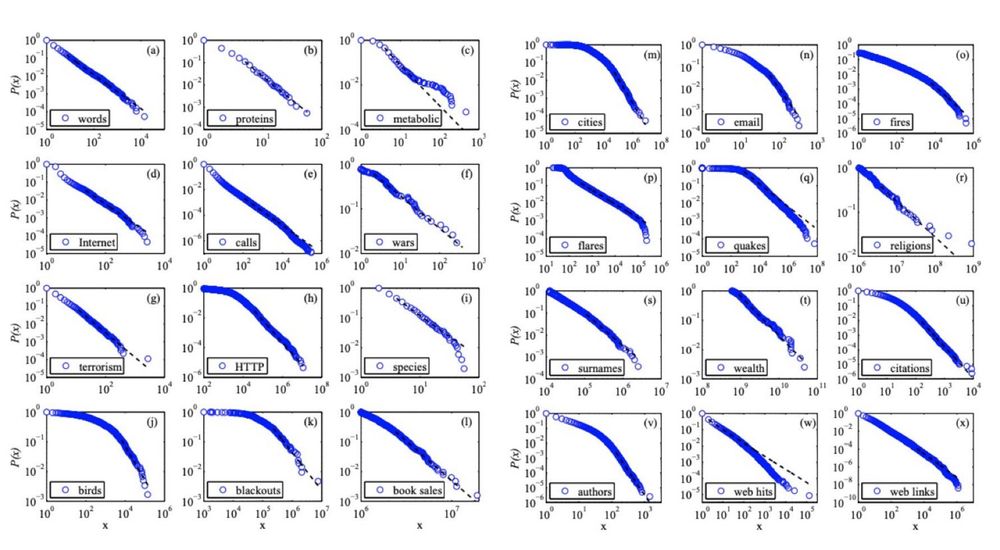
-Physicist Fritz Houtermans
There's a lot of truth to this. log-log plots are often abused and can be very misleading
1/5

Computational protein design starter pack. Let me know if I missed you!
Computational protein design starter pack. Let me know if I missed you!
Also fun, I remember being in the room for Ilya's talk of this paper (right after our talk on How transferable are features in deep neural networks?").
We got this idea after their cool work on improving Plug and Play with FM: arxiv.org/abs/2410.02423
I use it a lot for my protein design workflows together with @biotite.bsky.social.
Just `pip install pymol-remote`
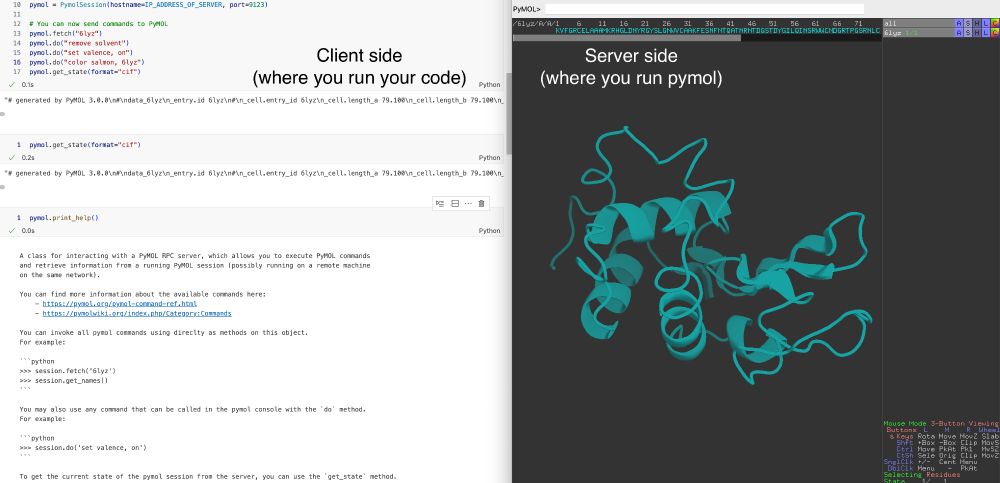
I use it a lot for my protein design workflows together with @biotite.bsky.social.
Just `pip install pymol-remote`
(In brief: Use ESM-2 650M and calculate mean embeddings across sites.)
www.biorxiv.org/content/10.1...

(In brief: Use ESM-2 650M and calculate mean embeddings across sites.)
www.biorxiv.org/content/10.1...
- thermophilic enzymes are not slower than mesophilic!
- general kcat/KM predictors are bad and easily beaten by models trained on this specific dataset!
www.biorxiv.org/content/10.1...

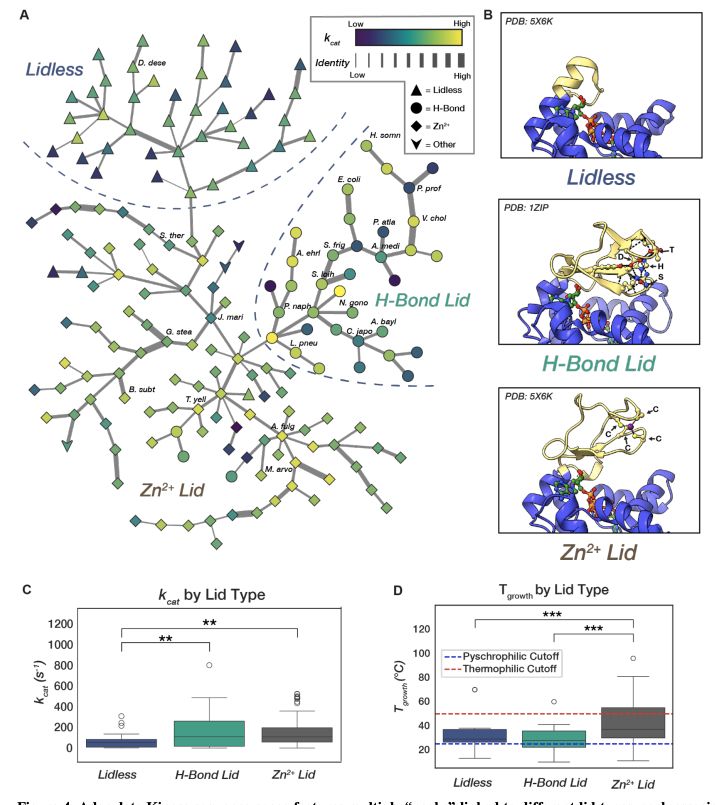

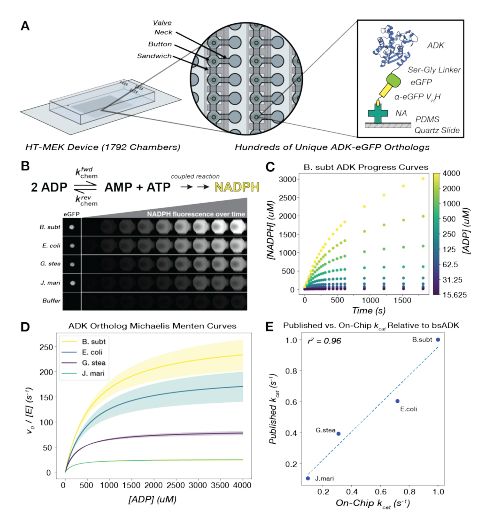
- thermophilic enzymes are not slower than mesophilic!
- general kcat/KM predictors are bad and easily beaten by models trained on this specific dataset!
www.biorxiv.org/content/10.1...



