mirdita.org

mirdita.org


youtu.be/7XnROkjo5Vg?...

•commented on biases in evolutionary signals from Tree of life used to train pLMs (a favorite paper I read in 2024: shorturl.at/fbC7g)

•commented on biases in evolutionary signals from Tree of life used to train pLMs (a favorite paper I read in 2024: shorturl.at/fbC7g)
Really proud of @amyxlu.bsky.social 's effort leading this project end-to-end!

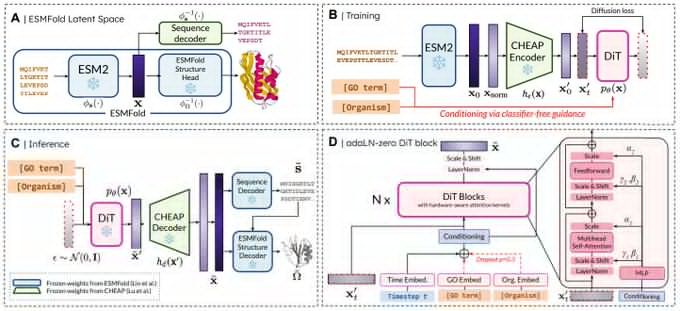

Really proud of @amyxlu.bsky.social 's effort leading this project end-to-end!
bit.ly/plaid-proteins
🧵
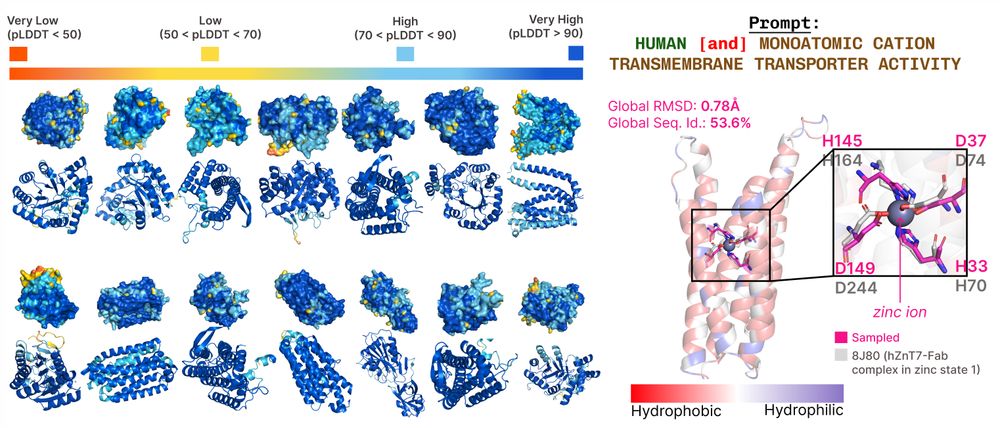
bit.ly/plaid-proteins
🧵

