

How can you transfer peptide IDs between runs and still control your false discovery rate? Till now, the short answer is, you couldn't. Now you can, with PIP-ECHO.
www.biorxiv.org/content/10.1...

How can you transfer peptide IDs between runs and still control your false discovery rate? Till now, the short answer is, you couldn't. Now you can, with PIP-ECHO.
www.biorxiv.org/content/10.1...
The buffering is much stronger than at the RNA level.

The buffering is much stronger than at the RNA level.
#proteomics #bioinformatics
#proteomics #bioinformatics
go.bsky.app/PUwu7fr
go.bsky.app/PUwu7fr

It explores how normalisation, the CV formula and software parameters affect the outputs. It suggests parameters to use for biological and technical studies and provides an R package to calculate CVs.
doi.org/10.1021/acs....
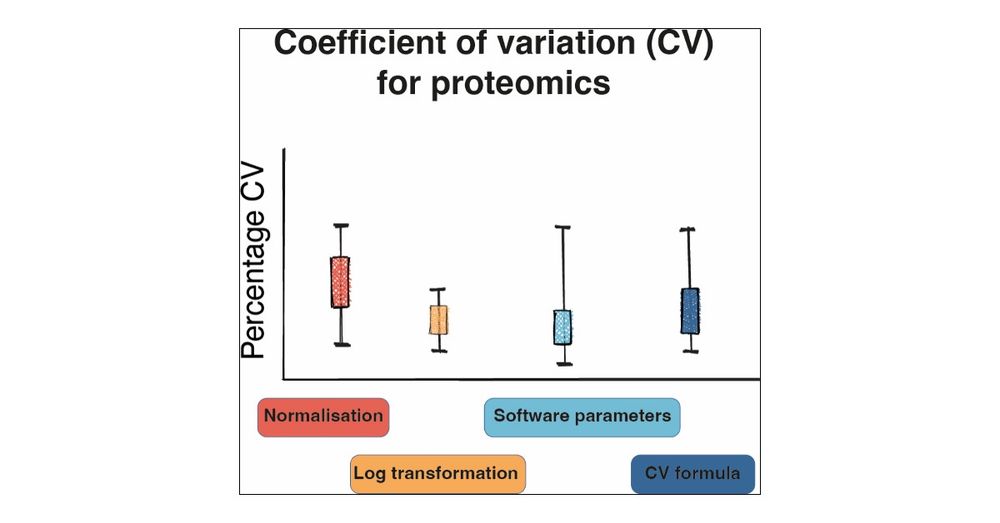
It explores how normalisation, the CV formula and software parameters affect the outputs. It suggests parameters to use for biological and technical studies and provides an R package to calculate CVs.
doi.org/10.1021/acs....




