
We found that by using ab initio reconstruction at very high res, in very small steps, we could crack some small structures that had eluded us - e.g. 39kDa iPKAc (EMPIAR-10252), below.
Read on for details... 1/x
We found that by using ab initio reconstruction at very high res, in very small steps, we could crack some small structures that had eluded us - e.g. 39kDa iPKAc (EMPIAR-10252), below.
Read on for details... 1/x
biorxiv.org/content/10.110
biorxiv.org/content/10.110
www.nature.com/articles/s41...
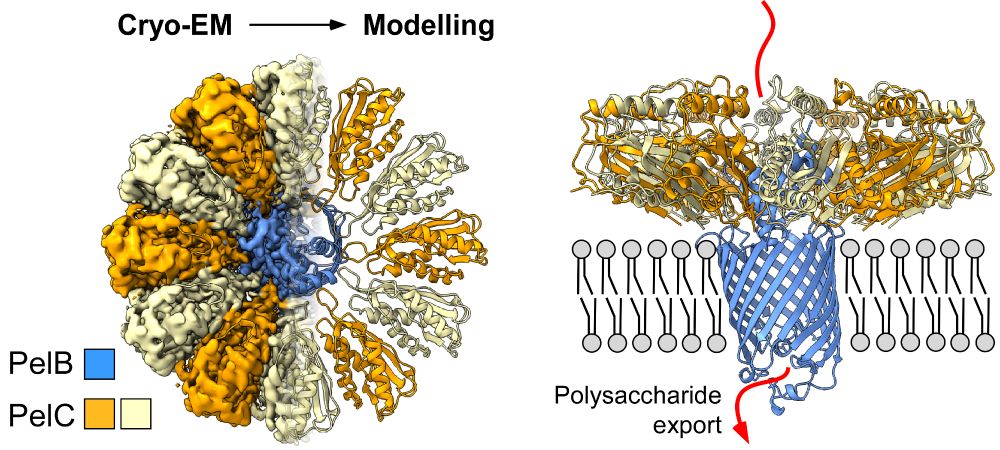
www.nature.com/articles/s41...
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...

Preprint📜: www.biorxiv.org/content/10.1...
A short thread🧵👇
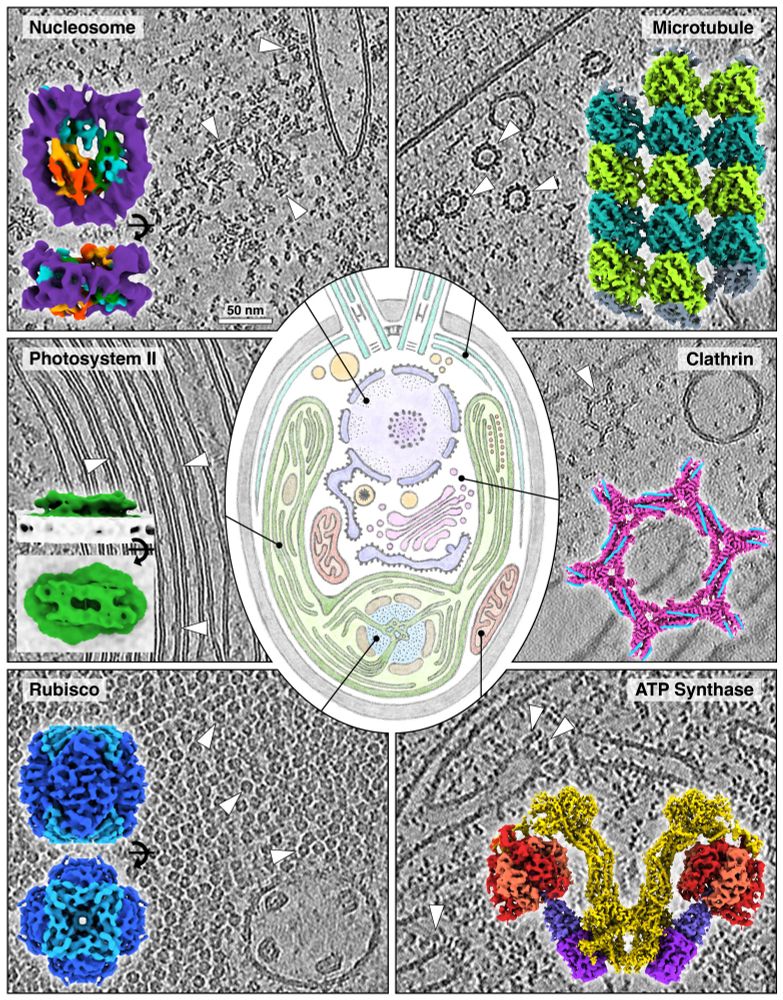
Preprint📜: www.biorxiv.org/content/10.1...
A short thread🧵👇

💾 github.com/SBQ-1999/Cry...
💾 github.com/SBQ-1999/Cry...
Feel free to share, add and distribute, so we can come together again! 😍
go.bsky.app/AEeXh86
Feel free to share, add and distribute, so we can come together again! 😍
go.bsky.app/AEeXh86

