
https://scholar.google.com/citations?user=K8qTnLUAAA

⏳ Closing September 16th.
✏️ Apply here: tinyurl.com/2ukkp8yx
Or learn more about what we do at www.lannelongue-group.org
Initial examples of research projects below 👇
⏳ Closing September 16th.
✏️ Apply here: tinyurl.com/2ukkp8yx
Or learn more about what we do at www.lannelongue-group.org
Initial examples of research projects below 👇
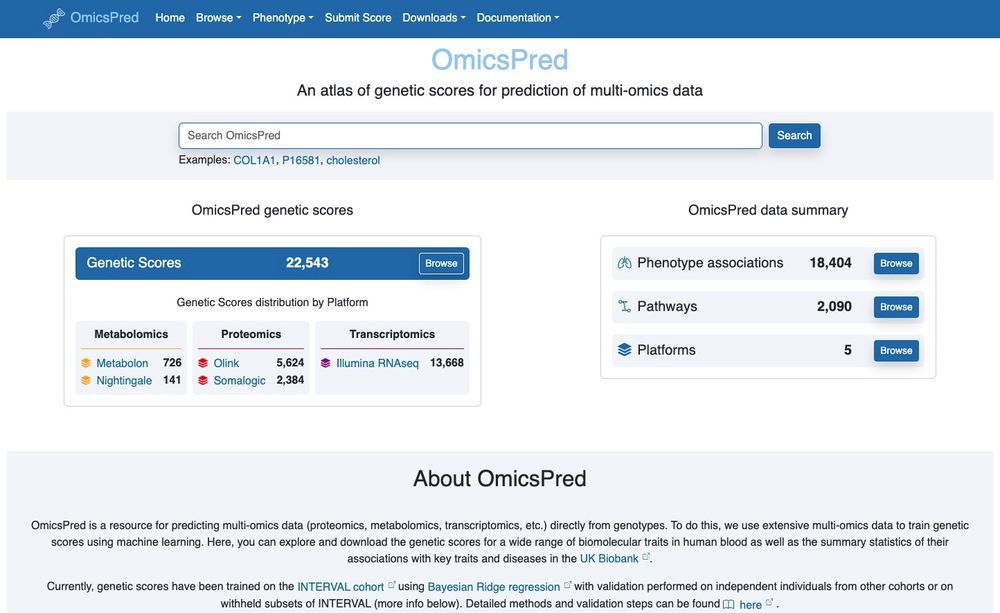
We've annotated/deposited the GTExV8 gene expression predictors so they are now available alongside all the other multi-omic predictors at www.omicspred.org
More on its way... a wonderful collab with @hakyim.bsky.social & co!
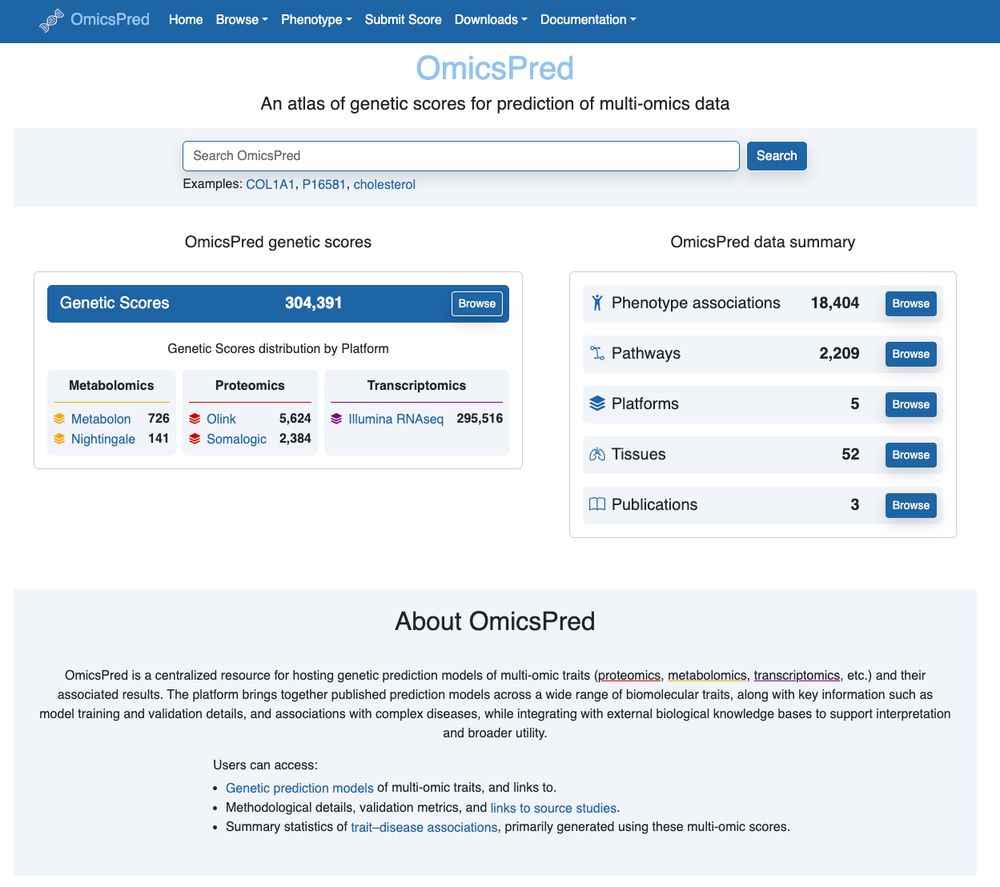
We've annotated/deposited the GTExV8 gene expression predictors so they are now available alongside all the other multi-omic predictors at www.omicspred.org
More on its way... a wonderful collab with @hakyim.bsky.social & co!
www.embopress.org/doi/full/10....
A short thread on our key findings 🧵👇

www.embopress.org/doi/full/10....
A short thread on our key findings 🧵👇
Check out the INTERVAL RNAseq portal www.intervalrna.org.uk
Led by @alextokolyi.bsky.social & Elodie Persyn!
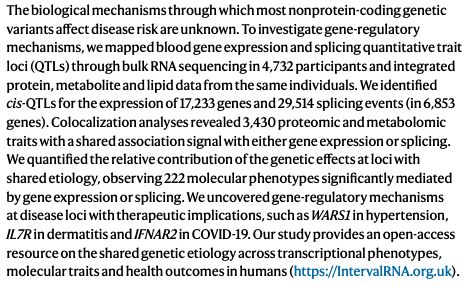
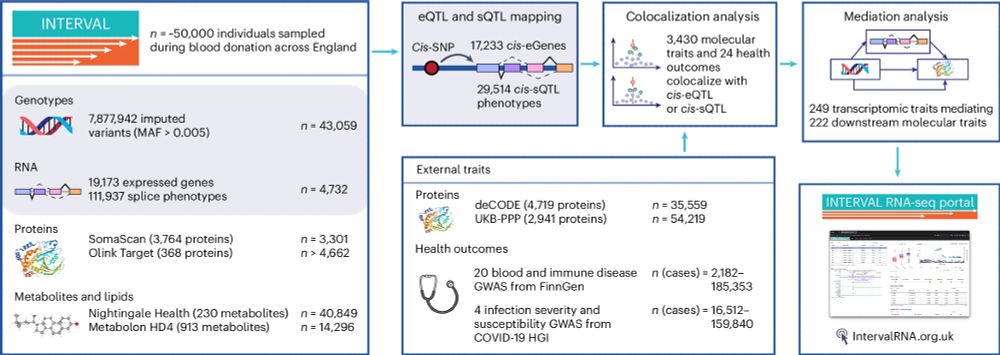
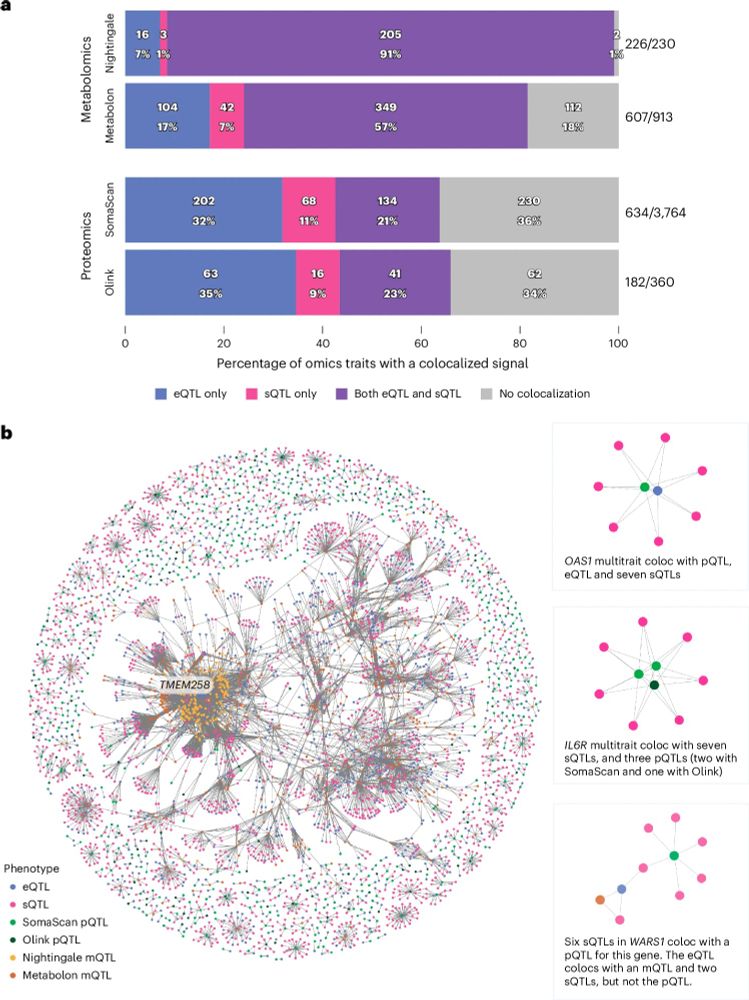
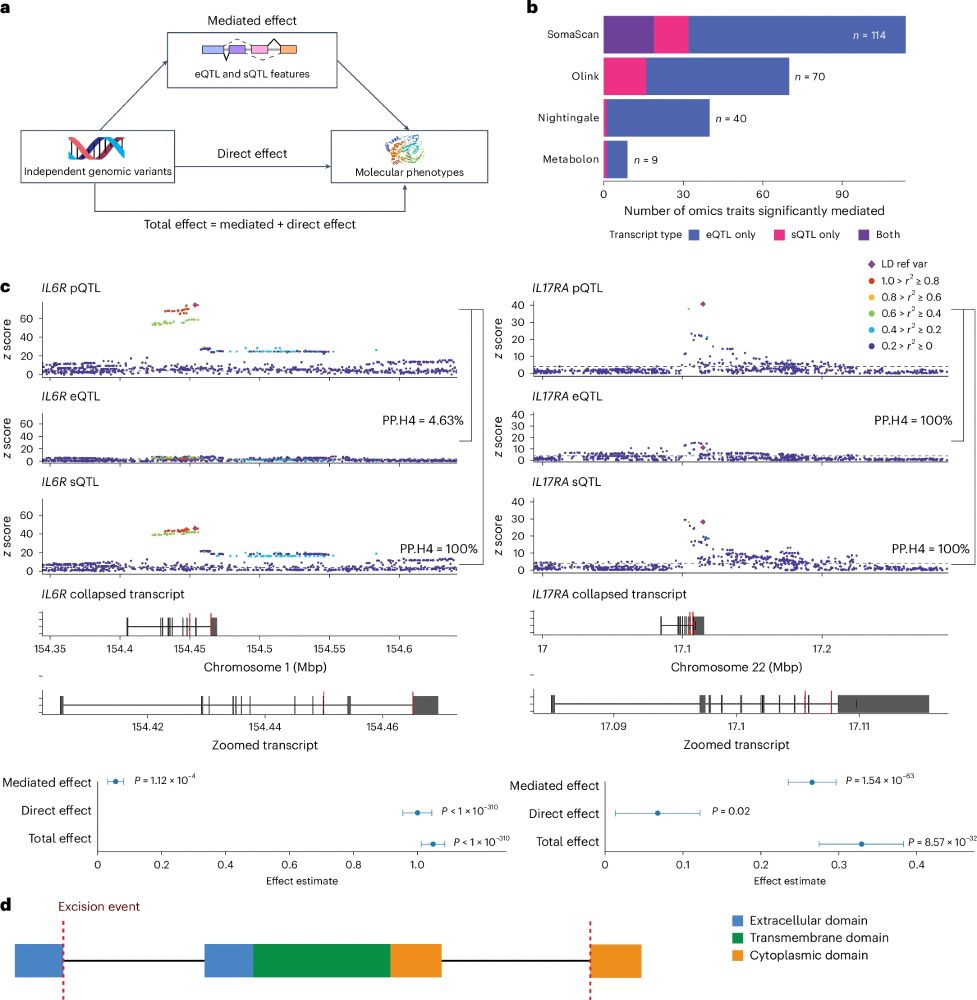
Check out the INTERVAL RNAseq portal www.intervalrna.org.uk
Led by @alextokolyi.bsky.social & Elodie Persyn!
Well done Doug Loesch, Dirk Paul, Abhishek Nag and co!




Well done Doug Loesch, Dirk Paul, Abhishek Nag and co!

Comparative Analysis Between Olink-PEA and Alamar-NULISA Proteomic Technologies Applied to a Critically Ill COVID-19 Cohort
analyticalsciencejournals.onlinelibrary.wiley.com/doi/10.1002/...

Comparative Analysis Between Olink-PEA and Alamar-NULISA Proteomic Technologies Applied to a Critically Ill COVID-19 Cohort
analyticalsciencejournals.onlinelibrary.wiley.com/doi/10.1002/...

A clinical consensus statement of the ESC Council on Cardiovascular Genomics, ESC Cardiovascular Risk Collaboration and European Association of Preventive Cardiology

A clinical consensus statement of the ESC Council on Cardiovascular Genomics, ESC Cardiovascular Risk Collaboration and European Association of Preventive Cardiology

Apply by March 7th at lpshg.com

Apply by March 7th at lpshg.com
🌱 More details and links to all adverts there: www.green-algorithms.org/join-us/
Closing 25/11
#AcademicSky #SciSky
Very excited to build a research group at Cambridge Uni to expand our Green Algorithms work thanks to the support of the Wellcome Trust
www.green-algorithms.org/join-us
#AcademicSky #SciSky
🌱 More details and links to all adverts there: www.green-algorithms.org/join-us/
Closing 25/11
#AcademicSky #SciSky

This is a major update that makes the package compatible with the UK Biobank Research Analysis Platform and shows removal of technical variation on the full NMR data release coming Jan 2025

This is a major update that makes the package compatible with the UK Biobank Research Analysis Platform and shows removal of technical variation on the full NMR data release coming Jan 2025
medrxiv.org/content/10.1...
Here, we show that NMR biomarker scores combined with PRSs and SCORE2 may have moderate population health benefits for 10-year CVD risk prediction prevention
Thread 1/3:
medrxiv.org/content/10.1...
Here, we show that NMR biomarker scores combined with PRSs and SCORE2 may have moderate population health benefits for 10-year CVD risk prediction prevention
Thread 1/3:

