https://singhlab.net

We harnessed a single-cell FM for decoding the long-debated relationship between genome arch. and gene coregulation. 1/
Preprint here: www.biorxiv.org/content/10.1...
Biological systems are inherently multi-scale. With the representational power and speed of PLMs, we can now bridge the molecular and systems scales, to study what makes each of us distinctive, immunity-wise.

Biological systems are inherently multi-scale. With the representational power and speed of PLMs, we can now bridge the molecular and systems scales, to study what makes each of us distinctive, immunity-wise.
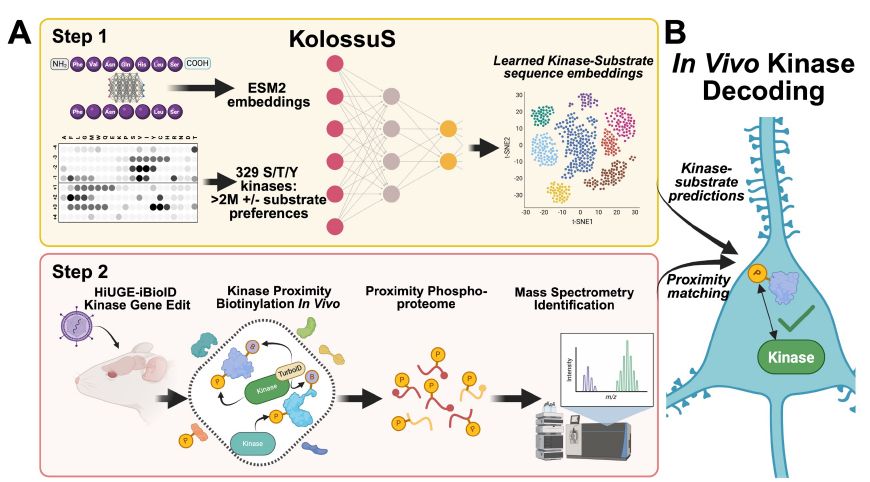
As computational biologists, our work mostly involves post-hoc analysis algorithms. KolossuS is the rare case where a ML model enables entirely new capabilities. 1/
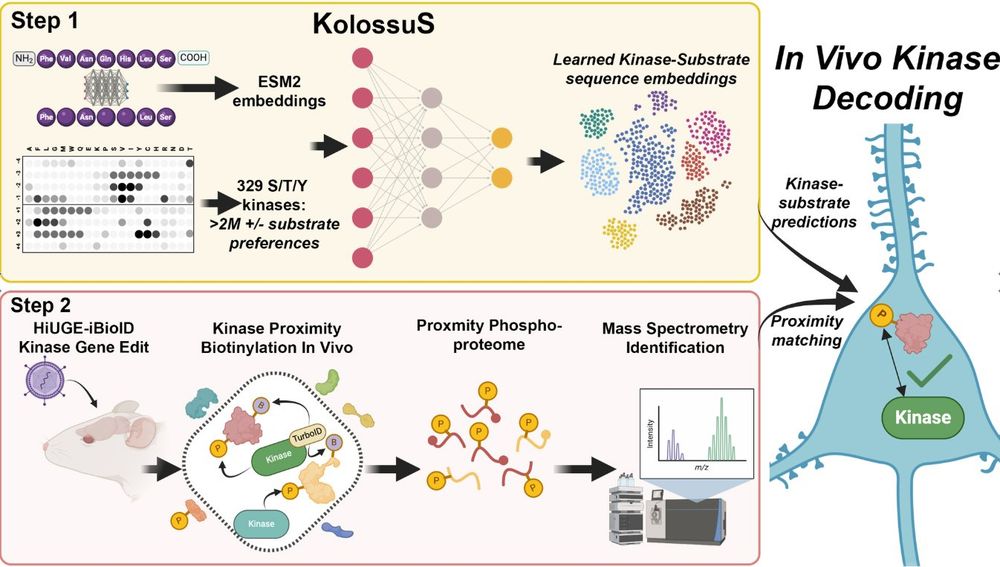
As computational biologists, our work mostly involves post-hoc analysis algorithms. KolossuS is the rare case where a ML model enables entirely new capabilities. 1/
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
@rohitsingh8080.bsky.social, DNA storage, and TAD inference




@rohitsingh8080.bsky.social, DNA storage, and TAD inference
We harnessed a single-cell FM for decoding the long-debated relationship between genome arch. and gene coregulation. 1/
Preprint here: www.biorxiv.org/content/10.1...

We harnessed a single-cell FM for decoding the long-debated relationship between genome arch. and gene coregulation. 1/
Preprint here: www.biorxiv.org/content/10.1...

We harnessed a single-cell FM for decoding the long-debated relationship between genome arch. and gene coregulation. 1/
Preprint here: www.biorxiv.org/content/10.1...
Explore the full paper: https://doi.org/10.1093/bioadv/vbaf060
#ProteinRepresentation #MachineLearning #Bioinformatics
@rohitsingh8080.bsky.social

Explore the full paper: https://doi.org/10.1093/bioadv/vbaf060
#ProteinRepresentation #MachineLearning #Bioinformatics
@rohitsingh8080.bsky.social

Explore the full paper: https://doi.org/10.1093/bioadv/vbaf060
#ProteinRepresentation #MachineLearning #Bioinformatics
@rohitsingh8080.bsky.social
www.cell.com/cell-systems...

www.cell.com/cell-systems...
All of this work is based on our framework generalizing Granger causality to directed acyclic graphs.
If you're at ENAR 2025, come say hi! We're recruiting! #ENAR2025
All of this work is based on our framework generalizing Granger causality to directed acyclic graphs.
If you're at ENAR 2025, come say hi! We're recruiting! #ENAR2025
Here's the preprint: www.biorxiv.org/content/10.1...

Here's the preprint: www.biorxiv.org/content/10.1...
The true measure of people's potential is not the height of the peak they've reached. It’s how far they've climbed to get there.
The true measure of people's potential is not the height of the peak they've reached. It’s how far they've climbed to get there.
Great talk on a variety of foods, from Higashi and SpiceMix to Steamboat!
Also, some stuff on 3D genome.

Great talk on a variety of foods, from Higashi and SpiceMix to Steamboat!
Also, some stuff on 3D genome.
Enter CoNCISE, our finite scalar quantization-based approach to identifying small molecule binders. 1/

Enter CoNCISE, our finite scalar quantization-based approach to identifying small molecule binders. 1/
This was my analysis of key hemoglobin-related genes.

This was my analysis of key hemoglobin-related genes.
A veritable zoo of PLMs now exists, so why should you care? Well, AbMAP reflects an opinionated design philosophy about antibody representation. And based on your needs, it may be a great fit! 1/
www.pnas.org/doi/10.1073/...
A veritable zoo of PLMs now exists, so why should you care? Well, AbMAP reflects an opinionated design philosophy about antibody representation. And based on your needs, it may be a great fit! 1/
www.pnas.org/doi/10.1073/...
A veritable zoo of PLMs now exists, so why should you care? Well, AbMAP reflects an opinionated design philosophy about antibody representation. And based on your needs, it may be a great fit! 1/
www.pnas.org/doi/10.1073/...
Trainees, come join us!
- I'm moving to Senior Applied Research Scientist in Digital Biology at NVIDIA (Jan '25)
- I'm starting a new lab at Duke as Visiting Assistant Prof (early '25)
Both roles focus on tackling hard problems in biological machine learning through collaborative research.
Long 🧵
Trainees, come join us!
explored the potential impact of computational protein design. Fantastic range of perspectives-- I was honored to contribute.
For all the tremendous translational excitement, I'd argue that the basic science potential may be even bigger.
explored the potential impact of computational protein design. Fantastic range of perspectives-- I was honored to contribute.
For all the tremendous translational excitement, I'd argue that the basic science potential may be even bigger.

