https://singhlab.net

As models go, KolossuS is relatively simple. In its design, we emphasized three aspects: breadth, calibration and interpretability. 7/

As models go, KolossuS is relatively simple. In its design, we emphasized three aspects: breadth, calibration and interpretability. 7/

As computational biologists, our work mostly involves post-hoc analysis algorithms. KolossuS is the rare case where a ML model enables entirely new capabilities. 1/
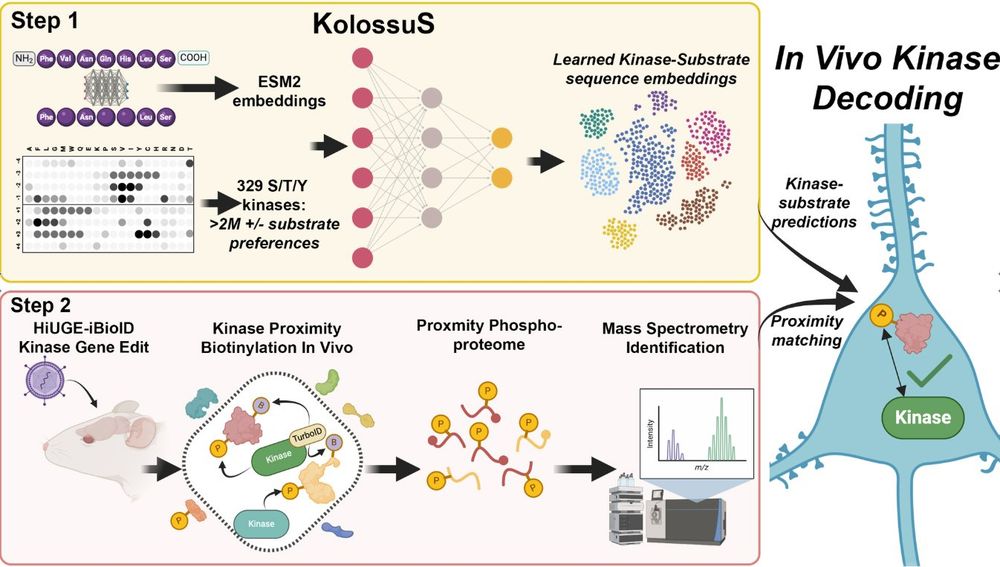
As computational biologists, our work mostly involves post-hoc analysis algorithms. KolossuS is the rare case where a ML model enables entirely new capabilities. 1/
In fact, we hypothesize TADs may work by synergizing with condensate formation. Check out the Perturb-seq analysis in our preprint. 12/

In fact, we hypothesize TADs may work by synergizing with condensate formation. Check out the Perturb-seq analysis in our preprint. 12/

Or has evolution encoded a synergy between chromatin conformation, gene location and gene function?
This is what we set out to explore quantitatively. 5/

Or has evolution encoded a synergy between chromatin conformation, gene location and gene function?
This is what we set out to explore quantitatively. 5/
Great talk on a variety of foods, from Higashi and SpiceMix to Steamboat!
Also, some stuff on 3D genome.

Great talk on a variety of foods, from Higashi and SpiceMix to Steamboat!
Also, some stuff on 3D genome.

The idea is that a 32d->32d->32d latent space is better organized than a flat 32,768-d latent space. 7/

The idea is that a 32d->32d->32d latent space is better organized than a flat 32,768-d latent space. 7/

Enter CoNCISE, our finite scalar quantization-based approach to identifying small molecule binders. 1/

Enter CoNCISE, our finite scalar quantization-based approach to identifying small molecule binders. 1/
One of my favorite figures in the paper (Fig 4B,C, Fig S1) show the striking AbMAP-space convergence between immune repertoires across nine individuals from
Bryan Briney's seminal study. 8/

One of my favorite figures in the paper (Fig 4B,C, Fig S1) show the striking AbMAP-space convergence between immune repertoires across nine individuals from
Bryan Briney's seminal study. 8/


