Feel free to reach out for advice.
www.nature.com/articles/s41...

Feel free to reach out for advice.
www.nature.com/articles/s41...

www.science.org/doi/10.1126/...

www.science.org/doi/10.1126/...
Our lab is searching for a technical assistant/research technician 🫀 to investigate heart regeneration and repair. Heidelberg is an awesome place to do science and the quality of living is great! Please forward to anyone who may be interested and please repost to spread the word 🙏

Our lab is searching for a technical assistant/research technician 🫀 to investigate heart regeneration and repair. Heidelberg is an awesome place to do science and the quality of living is great! Please forward to anyone who may be interested and please repost to spread the word 🙏
If you've ever wanted to dissect the subnuclear "neighborhood" around an individual locus, read on! (1/30)
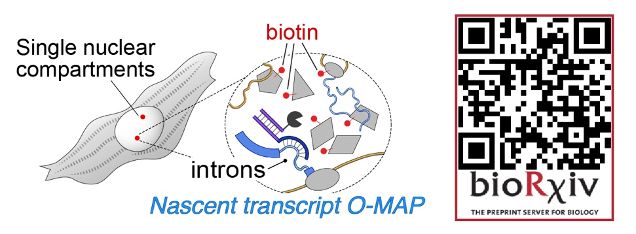

www.nature.com/articles/s41...

www.nature.com/articles/s41...
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...


