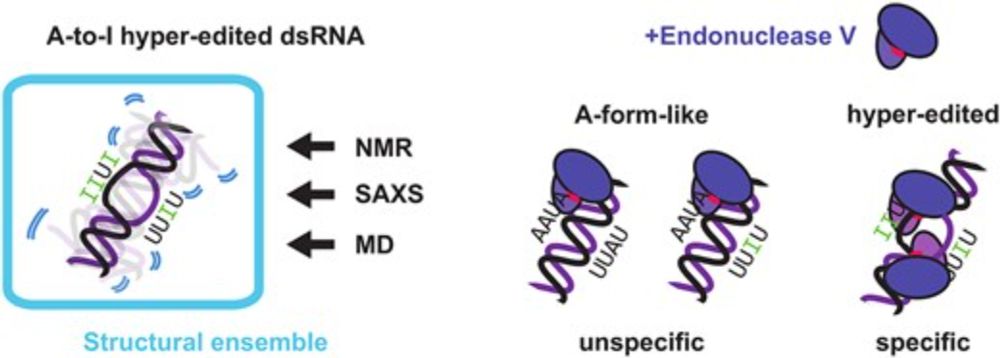
We expanded and refined the preprint thanks to the insightful feedback from reviewers!
paper: pubs.acs.org/doi/10.1021/...
code: github.com/RitAreaScien...

We expanded and refined the preprint thanks to the insightful feedback from reviewers!
paper: pubs.acs.org/doi/10.1021/...
code: github.com/RitAreaScien...

Special Guests: David Robertson (UK), @grovearmada.bsky.social (UK), Emanuele Andreano (IT)
www.areasciencepark.it/en/events/ai...

Special Guests: David Robertson (UK), @grovearmada.bsky.social (UK), Emanuele Andreano (IT)
www.areasciencepark.it/en/events/ai...
Join the Laboratory of Data Engineering to advance research in AI and its scientific applications.
We’re looking for motivated students ready to dive into interdisciplinary research in deep learning and AI.
Join the Laboratory of Data Engineering to advance research in AI and its scientific applications.
We’re looking for motivated students ready to dive into interdisciplinary research in deep learning and AI.

AlphaFold2: Protein conf. via evolution. Coevolution stabilizes.





