🔗 doi.org/10.1093/gbe/evaf091
#genome #evolution #introns
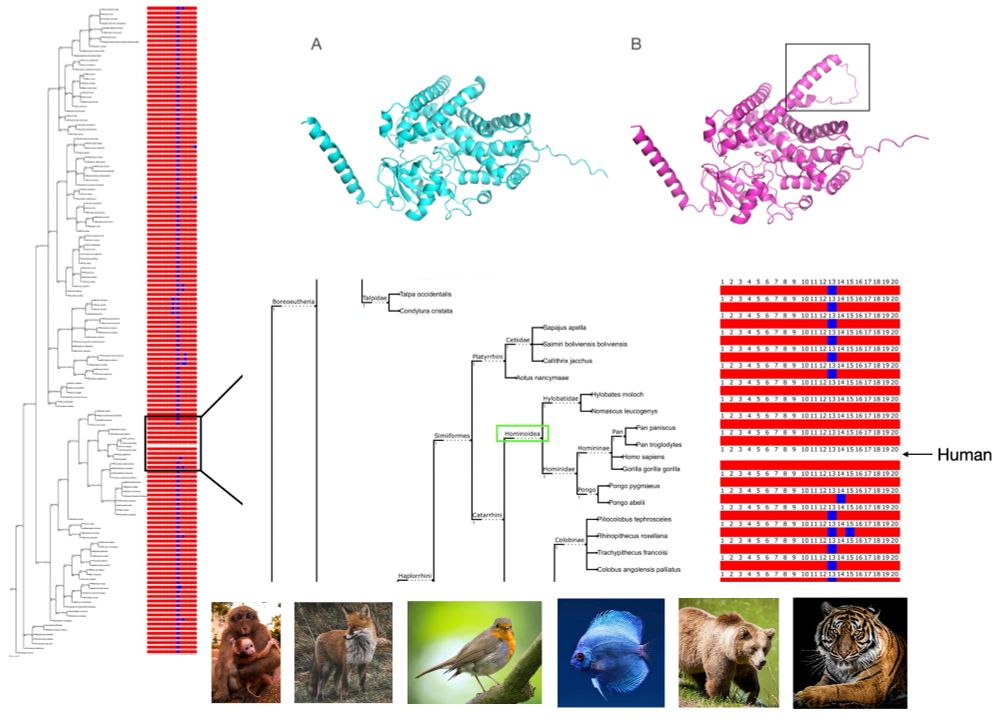
myloasm-docs.github.io
myloasm-docs.github.io
Mumemto scales to the new HPRC v2 release and beyond, and can merge in future assemblies without any recomputation! 1/n

Mumemto scales to the new HPRC v2 release and beyond, and can merge in future assemblies without any recomputation! 1/n



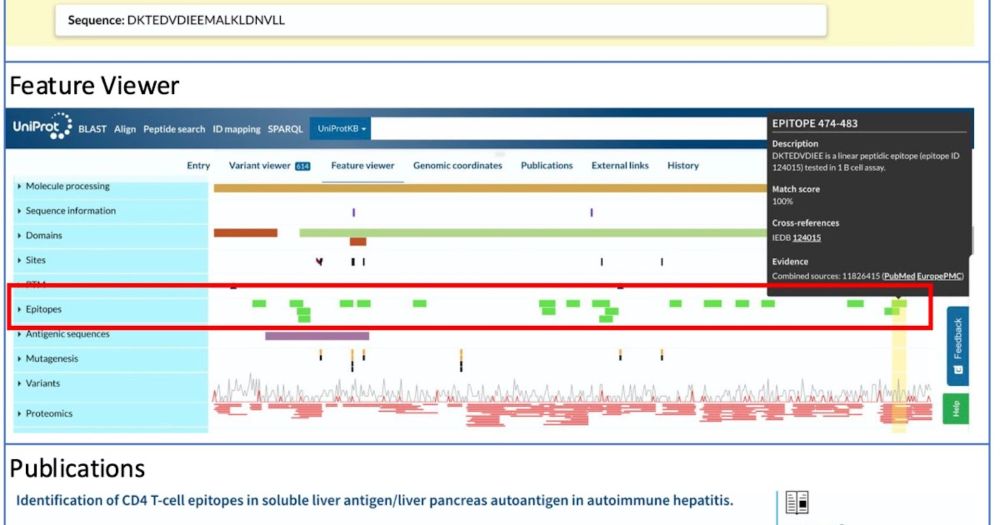
* abstract deadline: May 12 (AoE)
* paper deadline: May 15 (AoE)
Consider submitting your exciting algorithmic bioinformatics work to the WABI conference!
* abstract deadline: May 12 (AoE)
* paper deadline: May 15 (AoE)
Consider submitting your exciting algorithmic bioinformatics work to the WABI conference!

github.com/arcInstitute...

github.com/arcInstitute...
- We are seeking nominations bu June 1.
- September 18-19, 2025
- Please share with the star postdocs that you know.
docs.google.com/forms/d/e/1F...

- We are seeking nominations bu June 1.
- September 18-19, 2025
- Please share with the star postdocs that you know.
docs.google.com/forms/d/e/1F...
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...


- schematic and worked example for many algorithms
- alignment modes
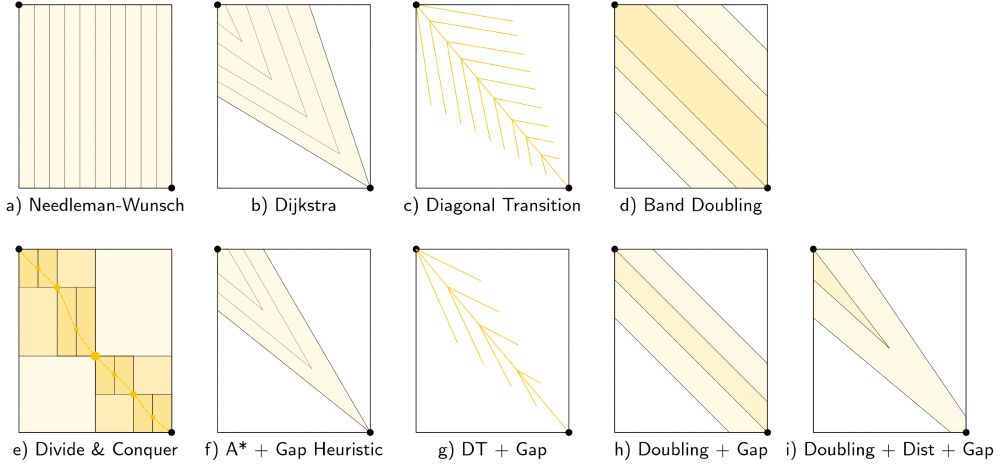
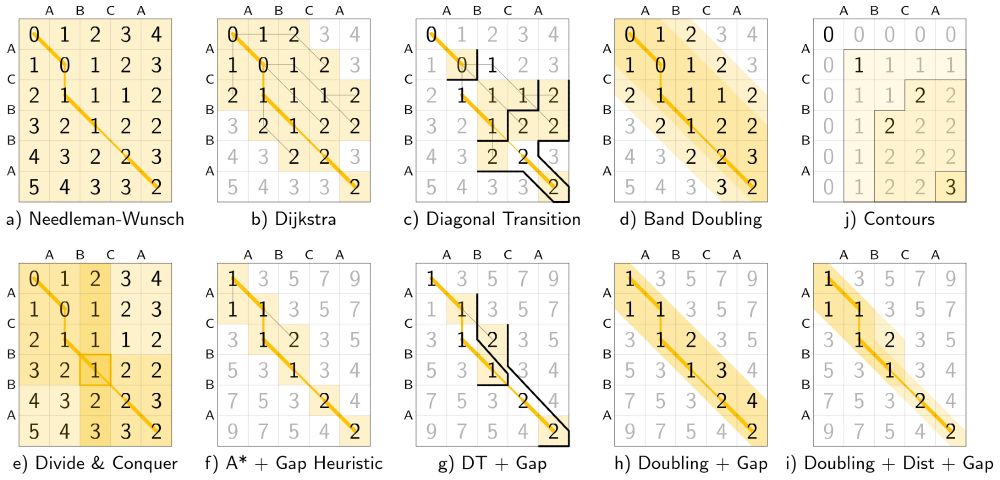
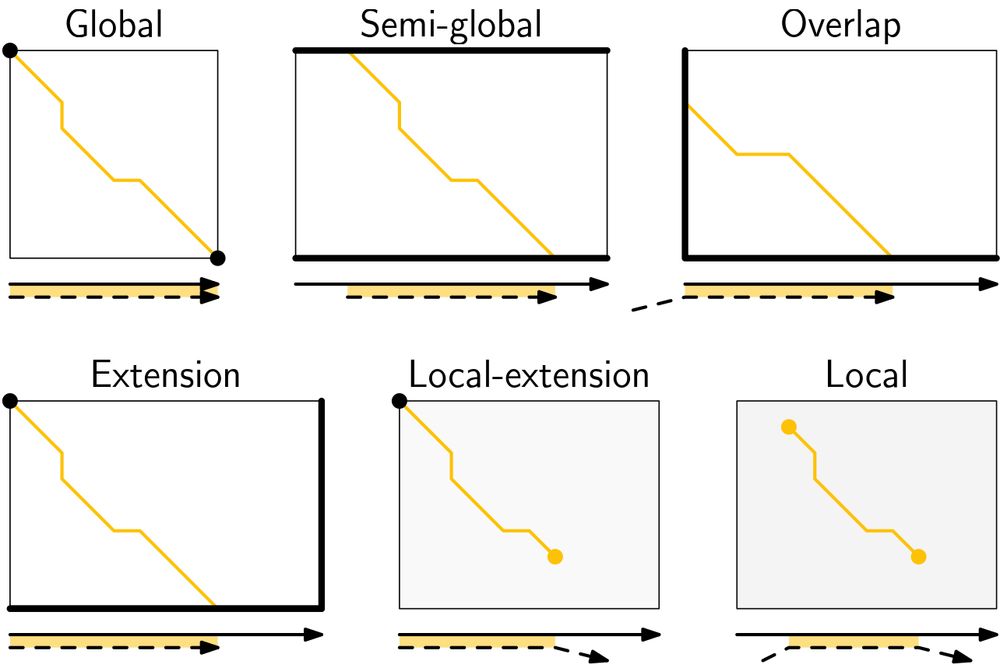
- schematic and worked example for many algorithms
- alignment modes
Thank-you to both Markus Schlegel from @activegroupgmbh.bsky.social for updating seq_io and Nicholas D. Crosbie of grepq for some competition and inspiration.
See more: github.com/fulcrumgenom...
Thank-you to both Markus Schlegel from @activegroupgmbh.bsky.social for updating seq_io and Nicholas D. Crosbie of grepq for some competition and inspiration.
See more: github.com/fulcrumgenom...

