
Bioinformatician working in Computational Proteomics.
More about me: https://michabirklbauer.me
Don’t miss your chance to join ProtAIomics and a vibrant international community in AI powered proteomics.
#ResearchCareers #Proteomics #ArtificialIntelligence #MSCA #PhDOpportunities #FullyFundedPhD

Don’t miss your chance to join ProtAIomics and a vibrant international community in AI powered proteomics.
#ResearchCareers #Proteomics #ArtificialIntelligence #MSCA #PhDOpportunities #FullyFundedPhD
www.nature.com/articles/s41...
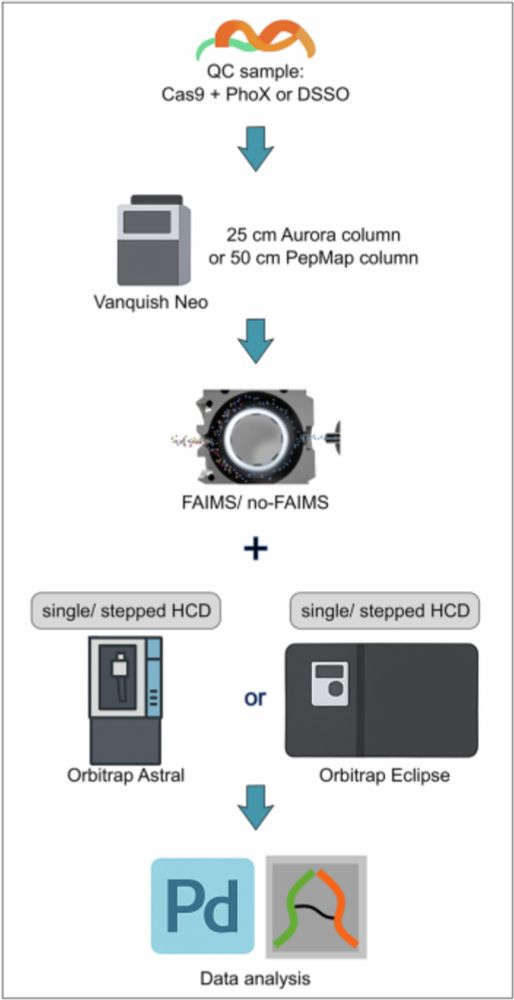
www.nature.com/articles/s41...

genzplyr is dplyr, but bussin fr fr no cap.

genzplyr is dplyr, but bussin fr fr no cap.
Join us in Harrachov, Czechia for a week of keynotes, workshops, and networking on computational MS.
Info and registration: eubic-ms.org/events/2026-...
#EuBIC2026 #MassSpectrometry #MassSpec #Bioinformatics #Proteomics #Metabolomics

Join us in Harrachov, Czechia for a week of keynotes, workshops, and networking on computational MS.
Info and registration: eubic-ms.org/events/2026-...
#EuBIC2026 #MassSpectrometry #MassSpec #Bioinformatics #Proteomics #Metabolomics

Enjoy these snapshots and make sure to sign up next year!
#viennabiocenter




Enjoy these snapshots and make sure to sign up next year!
#viennabiocenter

---
#proteomics #prot-paper

---
#proteomics #prot-paper
Info
ssp2025.squarespace.com

Info
ssp2025.squarespace.com
We are looking forward to interesting scientific contributions.

We are looking forward to interesting scientific contributions.

www.verwaltung.uni-halle.de/dezern3/Auss...
#Academicsky
#Chemsky
#teammassspec
www.verwaltung.uni-halle.de/dezern3/Auss...
#Academicsky
#Chemsky
#teammassspec


👉 Learn more about our open proteomics software benchmarking platform at proteobench.readthedocs.io.
#Proteomics #Bioinformatics #Benchmarking

👉 Learn more about our open proteomics software benchmarking platform at proteobench.readthedocs.io.
#Proteomics #Bioinformatics #Benchmarking
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
I’ve pushed a new version of lesSDRF to make high-throughput SDRF annotation faster and more intuitive.
Github issue: github.com/CompOmics/le...
Temporary app: refactorlessdrf.streamlit.app
I could use some feedback before making this the new lesSDRF 🙏
I’ve pushed a new version of lesSDRF to make high-throughput SDRF annotation faster and more intuitive.
Github issue: github.com/CompOmics/le...
Temporary app: refactorlessdrf.streamlit.app
I could use some feedback before making this the new lesSDRF 🙏
Next on the list submission of the package to 👉 @bioconductor.bsky.social
Next on the list submission of the package to 👉 @bioconductor.bsky.social

Today Altmetric is announcing a major development in collaboration with the Brain Occipital Lobe Learning Observation Centre Kalhausen-Strasbourg.
From today Altmetric will be able to determine what research paper you're thinking of, regardless of what you're actually writing/saying.
1/17
Today Altmetric is announcing a major development in collaboration with the Brain Occipital Lobe Learning Observation Centre Kalhausen-Strasbourg.
From today Altmetric will be able to determine what research paper you're thinking of, regardless of what you're actually writing/saying.
1/17

