Nanopore's getting accurate, but
1. Can this lead to better metagenome assemblies?
2. How, algorithmically, to leverage them?
with co-author Max Marin @mgmarin.bsky.social, supervised by Heng Li @lh3lh3.bsky.social
1 / N
Nanopore's getting accurate, but
1. Can this lead to better metagenome assemblies?
2. How, algorithmically, to leverage them?
with co-author Max Marin @mgmarin.bsky.social, supervised by Heng Li @lh3lh3.bsky.social
1 / N
❄️Winter School 2026 (12–23 Jan, Lausanne)
➡️Advanced methods in microbial community analysis
🧬Hands-on training in 16S, metagenomics, metatranscriptomics, functional annotation & ML
📍Free course, apply now: nccr-microbiomes.ch/education/january-short-course/
#NCCR #Microbiomes
❄️Winter School 2026 (12–23 Jan, Lausanne)
➡️Advanced methods in microbial community analysis
🧬Hands-on training in 16S, metagenomics, metatranscriptomics, functional annotation & ML
📍Free course, apply now: nccr-microbiomes.ch/education/january-short-course/
#NCCR #Microbiomes
Preprint 📝: biorxiv.org/content/10.1...
🧵 1/n
Preprint 📝: biorxiv.org/content/10.1...
🧵 1/n
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

This joint PhD project between the @rug.nl (Rampal Etienne & Martijn Egas) and @utrechtuniversity.bsky.social (Bram van Dijk) explores how simple prebiotic systems could evolve reliable information transfer.
Apply here: shorturl.at/WgYMF
myloasm-docs.github.io
myloasm-docs.github.io
This joint PhD project between the @rug.nl (Rampal Etienne & Martijn Egas) and @utrechtuniversity.bsky.social (Bram van Dijk) explores how simple prebiotic systems could evolve reliable information transfer.
Apply here: shorturl.at/WgYMF
This joint PhD project between the @rug.nl (Rampal Etienne & Martijn Egas) and @utrechtuniversity.bsky.social (Bram van Dijk) explores how simple prebiotic systems could evolve reliable information transfer.
Apply here: shorturl.at/WgYMF
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
doi.org/10.1038/s420...

doi.org/10.1038/s420...
Introducing pubScan, a new way to explore your PubMed publications through the lens of co-authorship.
🔍 Check it out at: pubscan.expressrna.org
❤️ Love it? Show your support by liking, sharing, and spreading the word.
#pubscan

Introducing pubScan, a new way to explore your PubMed publications through the lens of co-authorship.
🔍 Check it out at: pubscan.expressrna.org
❤️ Love it? Show your support by liking, sharing, and spreading the word.
#pubscan
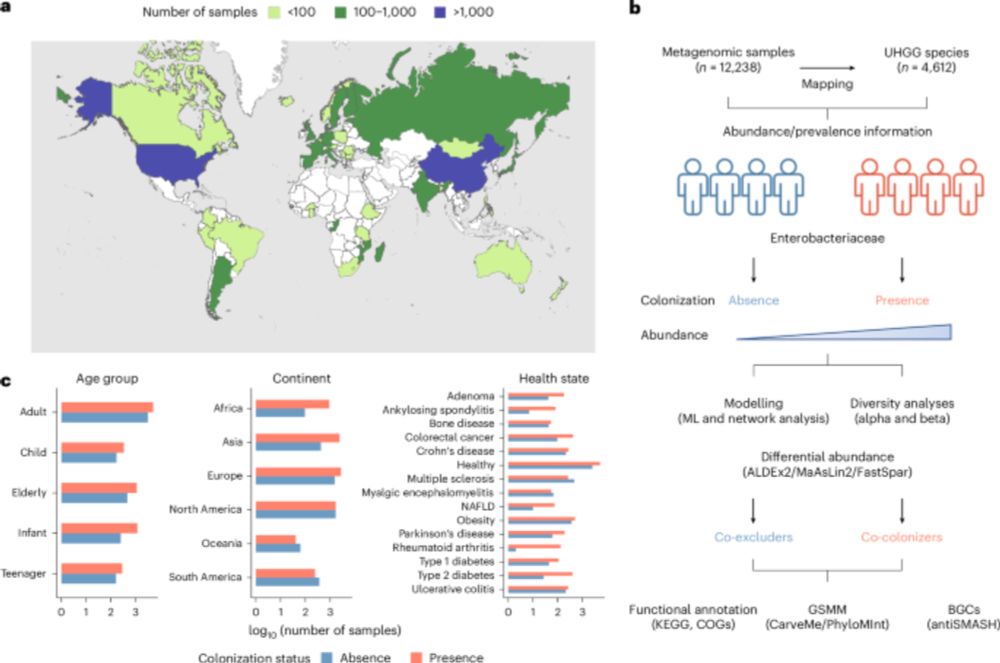
Here, we describe these emerging tools & their utility to both basic & translational microbiome research. @natmicrobiol.nature.com
www.nature.com/articles/s41...

Here, we describe these emerging tools & their utility to both basic & translational microbiome research. @natmicrobiol.nature.com
www.nature.com/articles/s41...

www.nature.com/articles/s41...
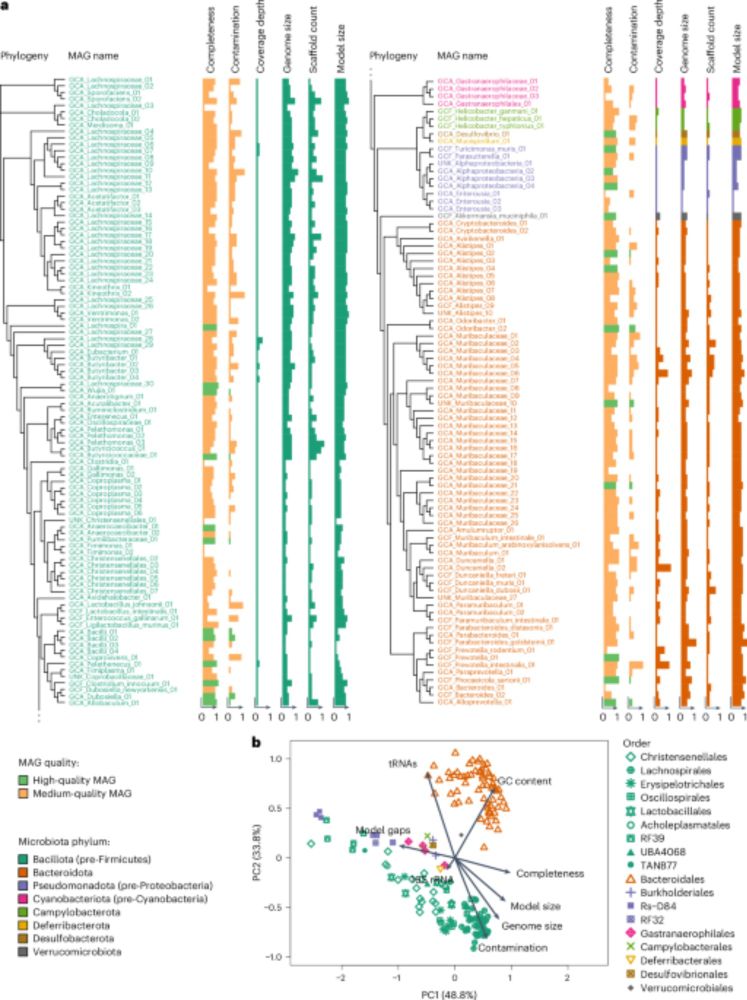
www.nature.com/articles/s41...

www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
📄 doi.org/10.1093/nsr/...
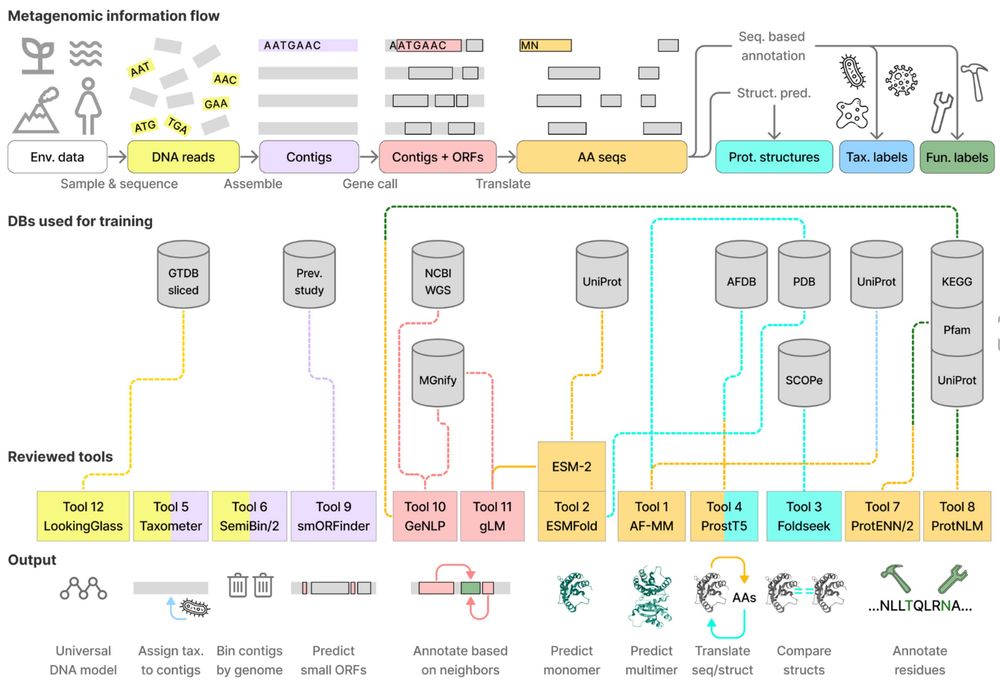
📄 doi.org/10.1093/nsr/...
Planning and describing a microbiome data analysis by @amydwillis.bsky.social and @davidandacat.bsky.social
www.nature.com/articles/s41...
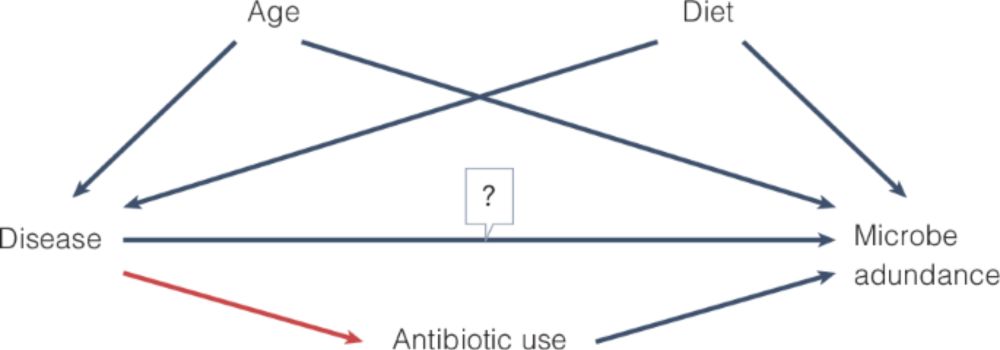
Planning and describing a microbiome data analysis by @amydwillis.bsky.social and @davidandacat.bsky.social
www.nature.com/articles/s41...
www.biorxiv.org/content/10.1...
micro-bites.org/author/ghada...
#soil_microbes ##SciComm
www.biorxiv.org/content/10.1...
micro-bites.org/author/ghada...
#soil_microbes ##SciComm
@kiranrpatil.bsky.social:
Obligate cross-feeding of metabolites is common in soil microbial communities
By Ghada Yousif @metagenomez.bsky.social with @swagatika.bsky.social @isamirgiri.bsky.social Sharvari Harshe et al.
www.biorxiv.org/content/10.1...
🧵👇

@kiranrpatil.bsky.social:
Obligate cross-feeding of metabolites is common in soil microbial communities
By Ghada Yousif @metagenomez.bsky.social with @swagatika.bsky.social @isamirgiri.bsky.social Sharvari Harshe et al.
www.biorxiv.org/content/10.1...
🧵👇

@kiranrpatil.bsky.social:
Obligate cross-feeding of metabolites is common in soil microbial communities
By Ghada Yousif @metagenomez.bsky.social with @swagatika.bsky.social @isamirgiri.bsky.social Sharvari Harshe et al.
www.biorxiv.org/content/10.1...
🧵👇