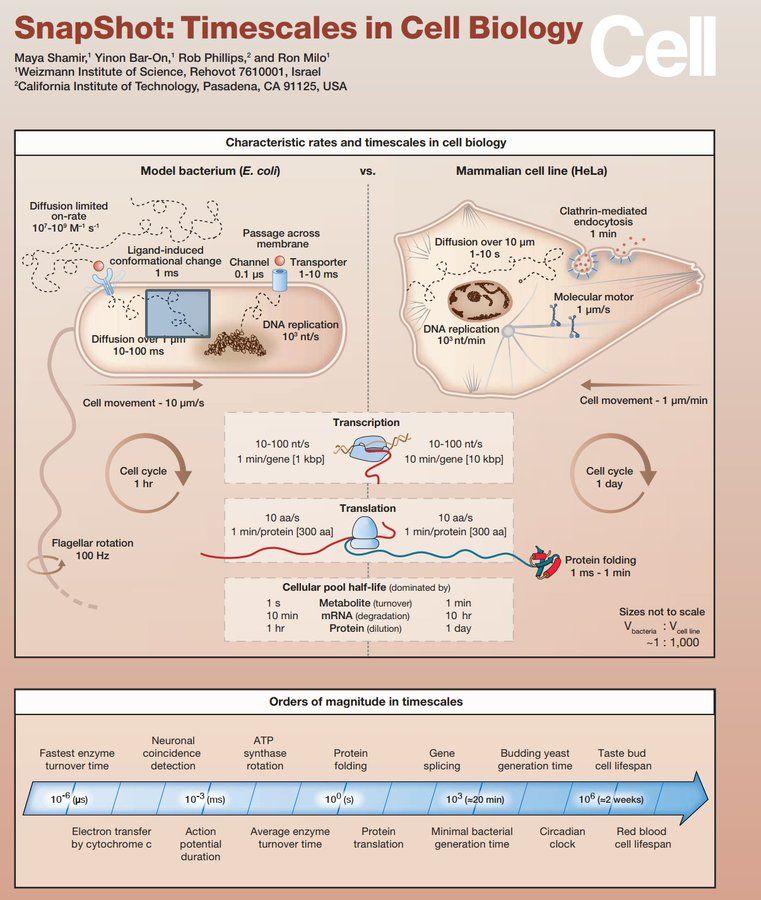
🔬🦠
docs.google.com/forms/d/e/1F...
🧫🧪 #ArchaeaSky #MicrobiomeSky

docs.google.com/forms/d/e/1F...
🧫🧪 #ArchaeaSky #MicrobiomeSky
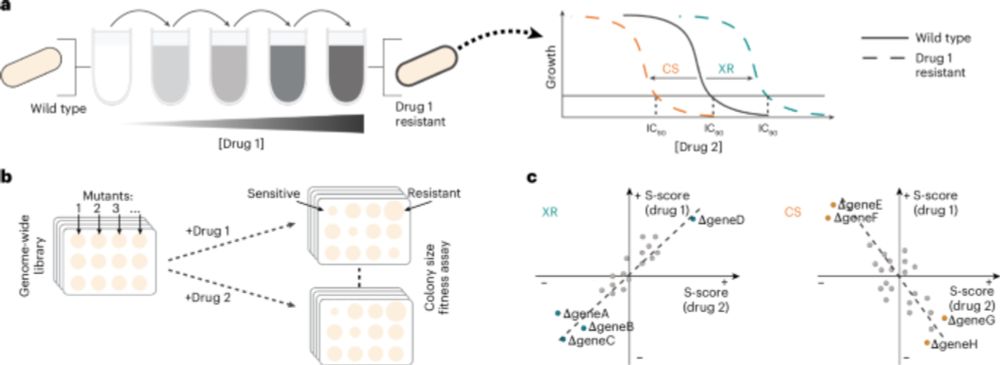
Dynamics of CRISPR-mediated virus-host interactions in the human gut microbiome
www.biorxiv.org/content/10.1...
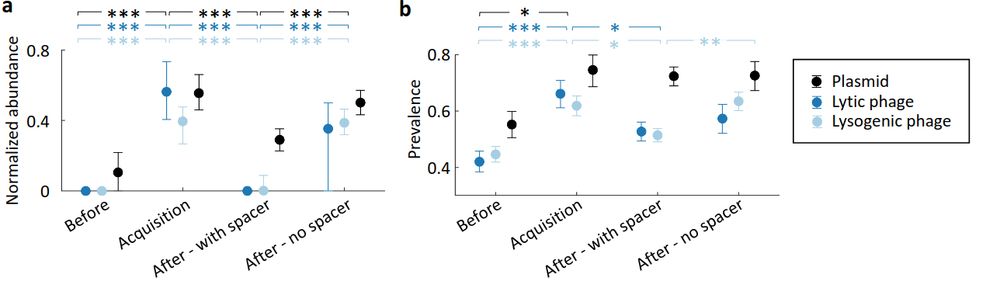
Dynamics of CRISPR-mediated virus-host interactions in the human gut microbiome
www.biorxiv.org/content/10.1...
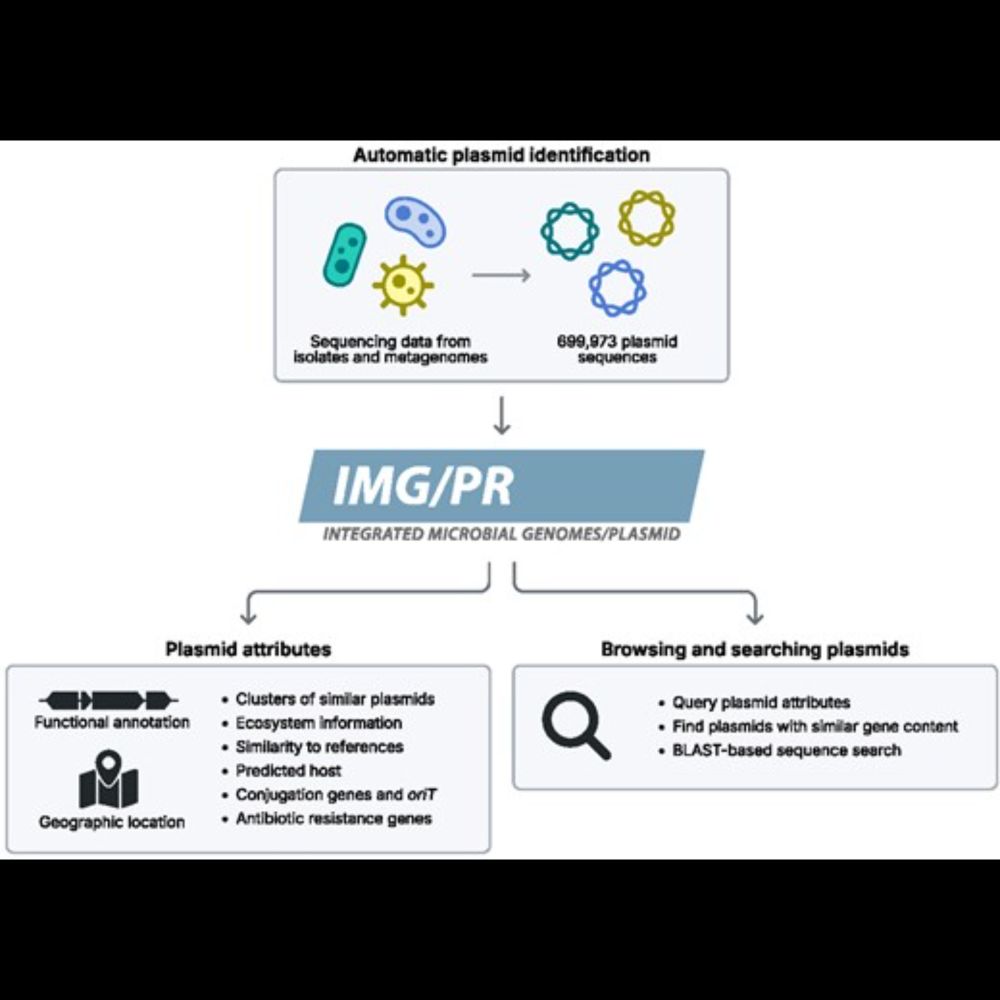
#DataViz protocols: An introduction to data visualization protocols for wet lab scientists#
A fantastic resource from @joachimgoedhart.bsky.social 🙏👏👍
joachimgoedhart.github.io/DataViz-prot...
#DataViz protocols: An introduction to data visualization protocols for wet lab scientists#
A fantastic resource from @joachimgoedhart.bsky.social 🙏👏👍
joachimgoedhart.github.io/DataViz-prot...

3 days of very interesting scientific talks and great conversation.
Shout-out to @jmarsh.bsky.social for being my biggest support and an awesome supervisor!
🦠



3 days of very interesting scientific talks and great conversation.
Shout-out to @jmarsh.bsky.social for being my biggest support and an awesome supervisor!
🦠




