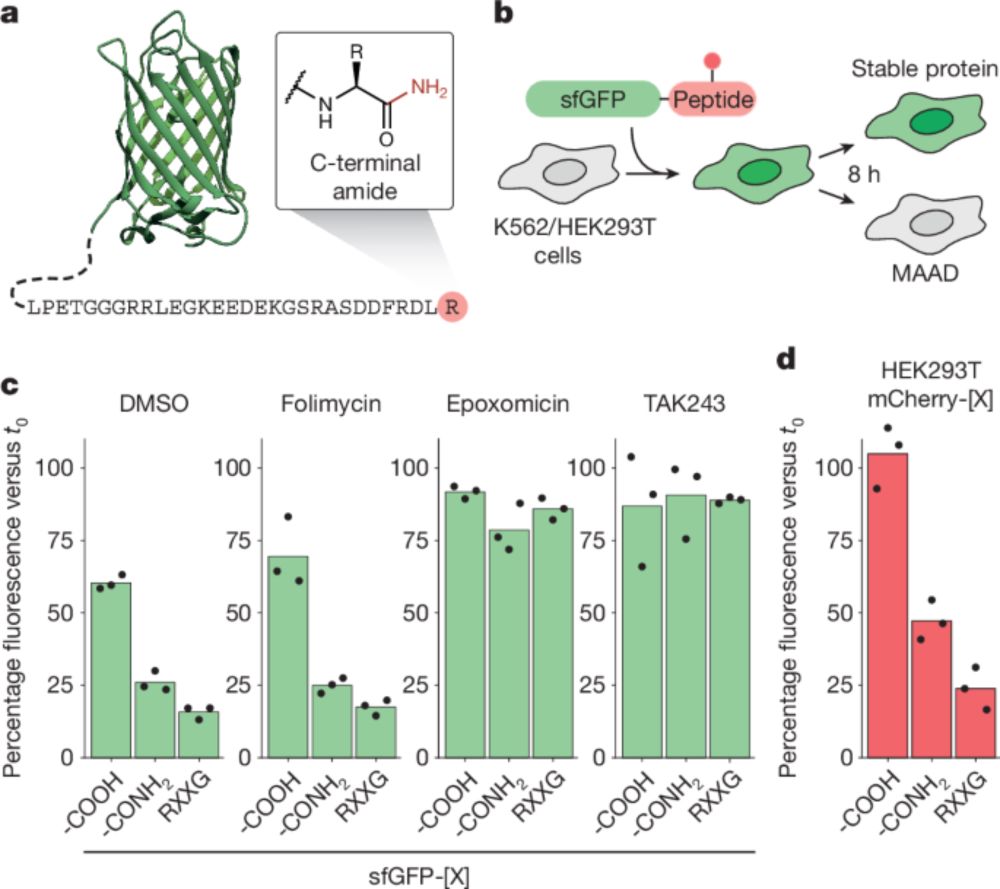
Super cool mechanism of how a metabolite regulates the stability of its own metabolizing enzyme!
Alina did it all for this project CRISPR screen ✂️, biochemistry 🧪, and cryo-EM ❄️🔬.
Congrats!
We are excited to share our recent work on #E3 ligase regulation in #metabolism!
www.biorxiv.org/content/10.6...
#ubiquitin #targetedproteindegradation #chemicalbiology
1/6
Super cool mechanism of how a metabolite regulates the stability of its own metabolizing enzyme!
Alina did it all for this project CRISPR screen ✂️, biochemistry 🧪, and cryo-EM ❄️🔬.
Congrats!



🔬🦠I'm hiring at all levels! 🔬🦠Check: www.molgen.mpg.de/fueyo-lab
🔬🦠I'm hiring at all levels! 🔬🦠Check: www.molgen.mpg.de/fueyo-lab

Ie: it's not us (Samtools team)! Be warned
Ie: it's not us (Samtools team)! Be warned


From the next application rounds, expect changes to the:
• proposal structure
• evaluation process
• extra funding you can request
• eligibility for Starting & Consolidator #Grants (from 2027)
More 👇 europa.eu/!RPHWvv

From the next application rounds, expect changes to the:
• proposal structure
• evaluation process
• extra funding you can request
• eligibility for Starting & Consolidator #Grants (from 2027)
More 👇 europa.eu/!RPHWvv
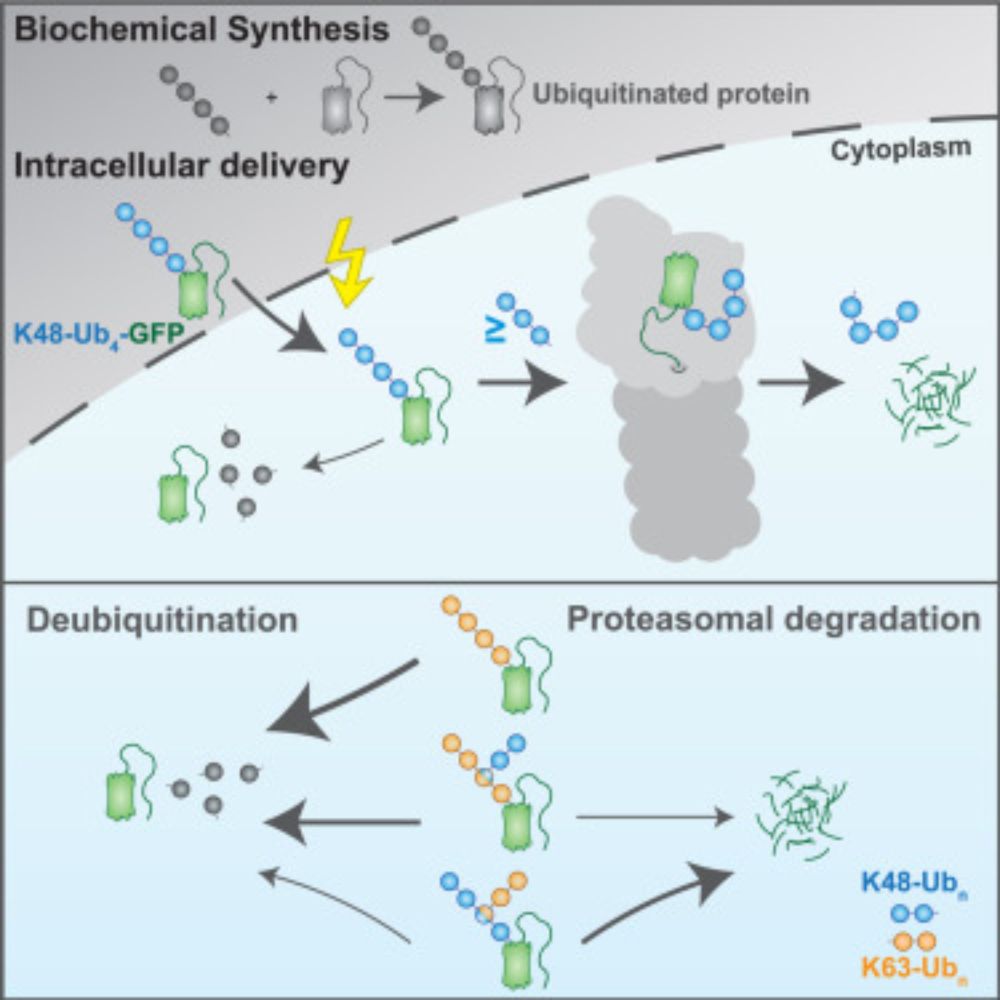
“𝗨𝗯𝗶𝗥𝗘𝗔𝗗 𝗱𝗲𝗰𝗶𝗽𝗵𝗲𝗿𝘀 𝗽𝗿𝗼𝘁𝗲𝗮𝘀𝗼𝗺𝗮𝗹 𝗱𝗲𝗴𝗿𝗮𝗱𝗮𝘁𝗶𝗼𝗻 𝗰𝗼𝗱𝗲 𝗼𝗳 𝗵𝗼𝗺𝗼𝘁𝘆𝗽𝗶𝗰 𝗮𝗻𝗱 𝗯𝗿𝗮𝗻𝗰𝗵𝗲𝗱 𝗞48 𝗮𝗻𝗱 𝗞63 𝘂𝗯𝗶𝗾𝘂𝗶𝘁𝗶𝗻 𝗰𝗵𝗮𝗶𝗻𝘀”
@cp-molcell.bsky.social
1/8
www.cell.com/molecular-ce...
“𝗨𝗯𝗶𝗥𝗘𝗔𝗗 𝗱𝗲𝗰𝗶𝗽𝗵𝗲𝗿𝘀 𝗽𝗿𝗼𝘁𝗲𝗮𝘀𝗼𝗺𝗮𝗹 𝗱𝗲𝗴𝗿𝗮𝗱𝗮𝘁𝗶𝗼𝗻 𝗰𝗼𝗱𝗲 𝗼𝗳 𝗵𝗼𝗺𝗼𝘁𝘆𝗽𝗶𝗰 𝗮𝗻𝗱 𝗯𝗿𝗮𝗻𝗰𝗵𝗲𝗱 𝗞48 𝗮𝗻𝗱 𝗞63 𝘂𝗯𝗶𝗾𝘂𝗶𝘁𝗶𝗻 𝗰𝗵𝗮𝗶𝗻𝘀”
@cp-molcell.bsky.social
1/8
www.cell.com/molecular-ce...