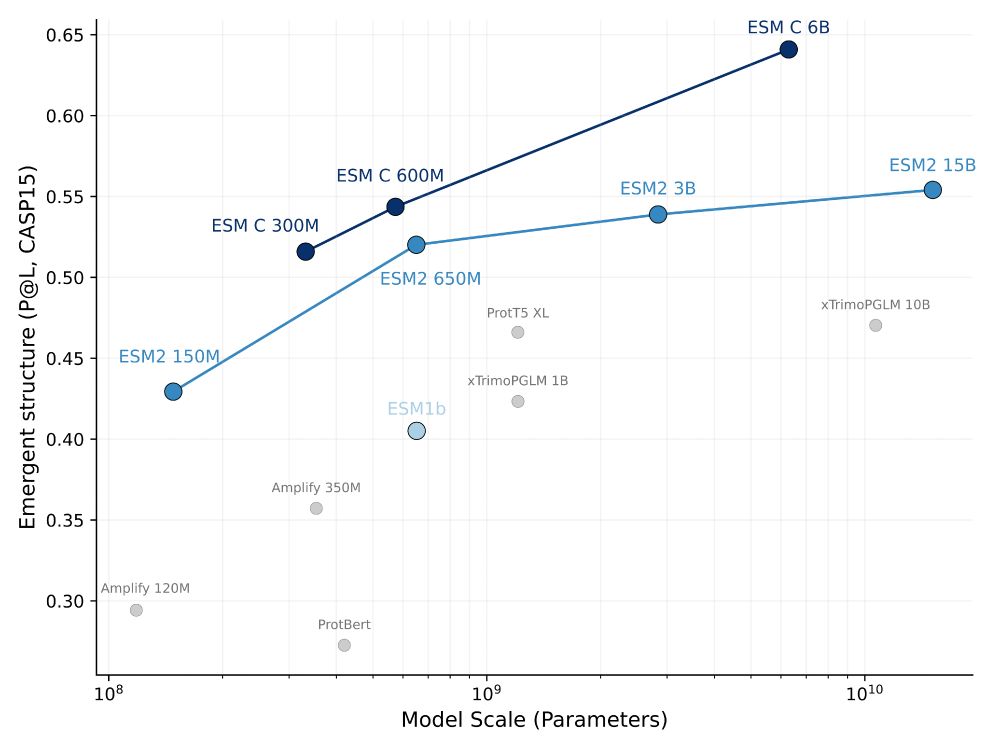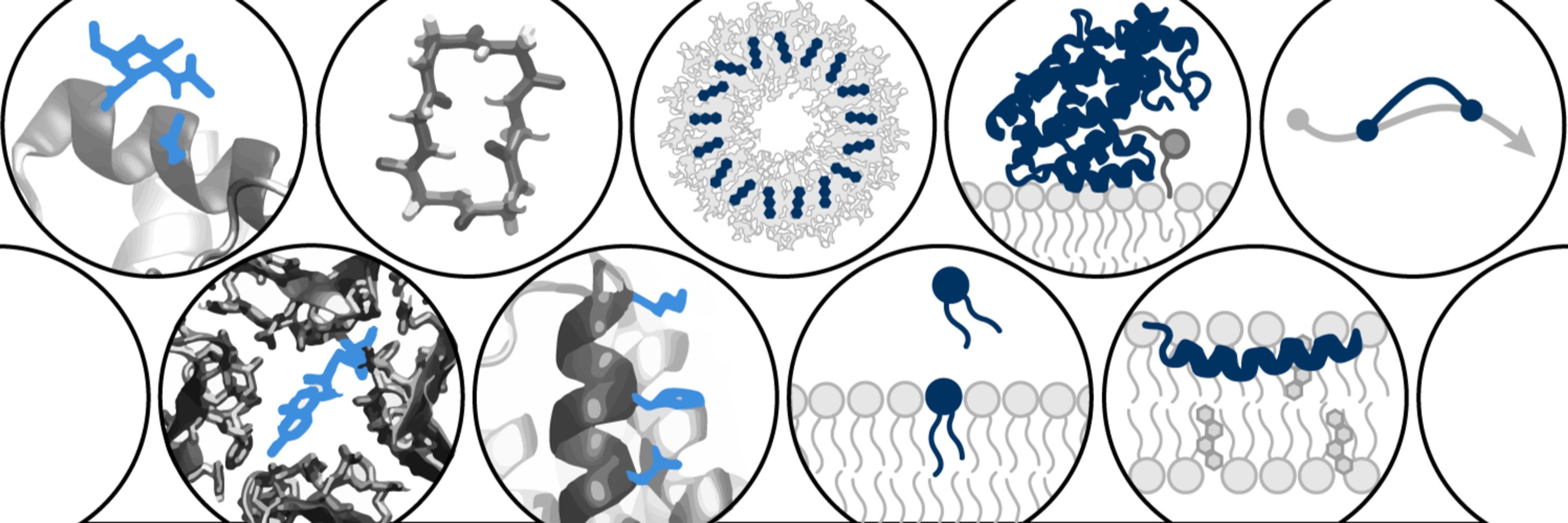

We have a great lineup of keynotes, contributed talks, and posters today and tomorrow
Schedule: mlcb.org/schedule
Join for free via livestream: m.youtube.com/@mlcbconf


🌐 afesm.foldseek.com
📄 www.biorxiv.org/content/10.1...

🌐 afesm.foldseek.com
📄 www.biorxiv.org/content/10.1...
We know proteins fluctuate between different conformations- but by how much? How does it vary from protein to protein? Can highly stable domains have low stability segments? @ajrferrari.bsky.social experimentally tested >5,000 domains to find out!
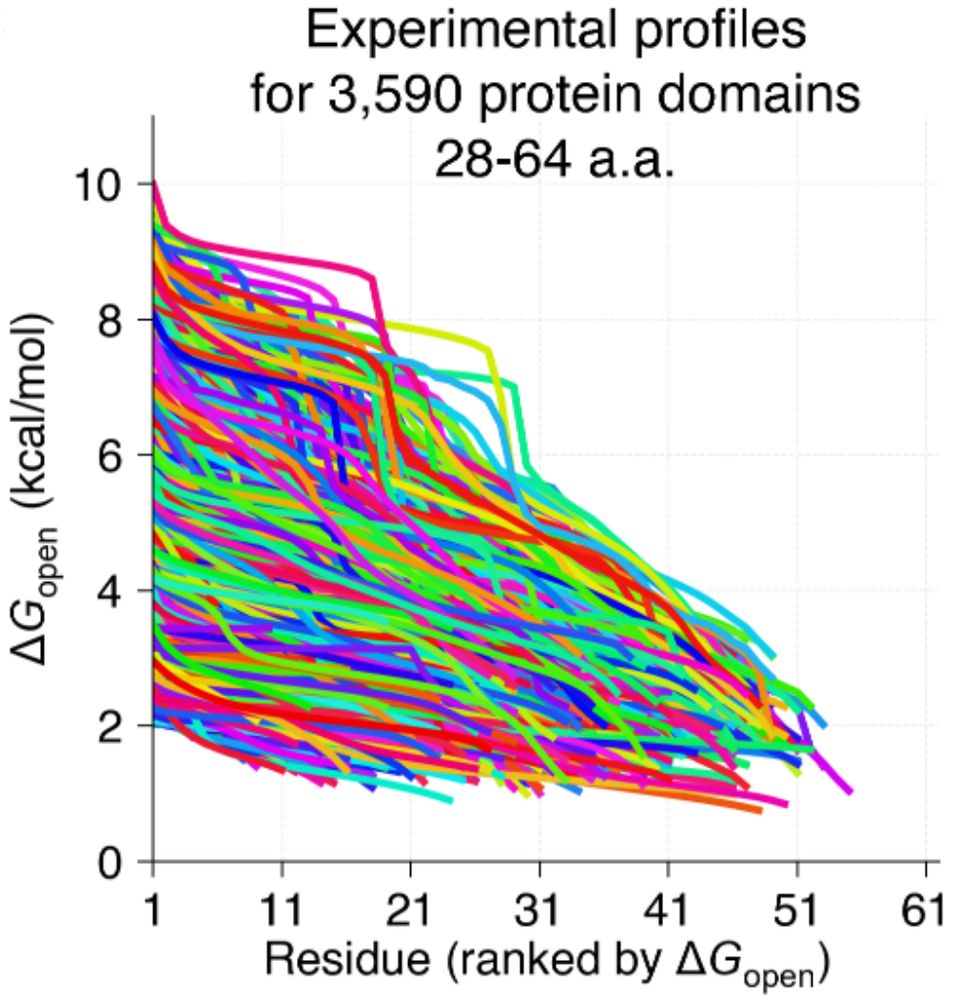
We know proteins fluctuate between different conformations- but by how much? How does it vary from protein to protein? Can highly stable domains have low stability segments? @ajrferrari.bsky.social experimentally tested >5,000 domains to find out!
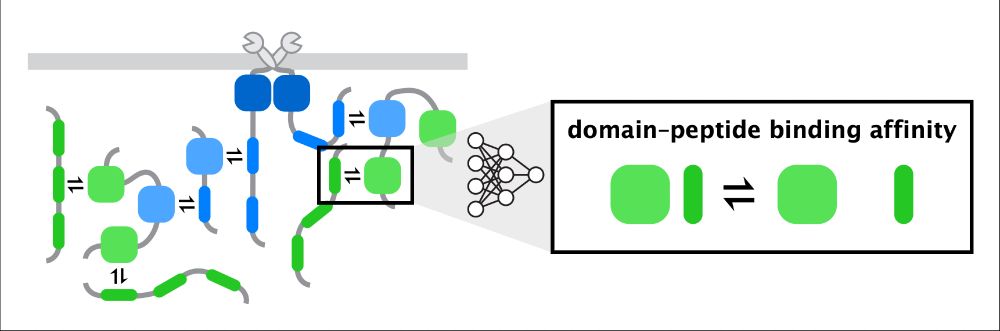
www.biorxiv.org/content/10.1...
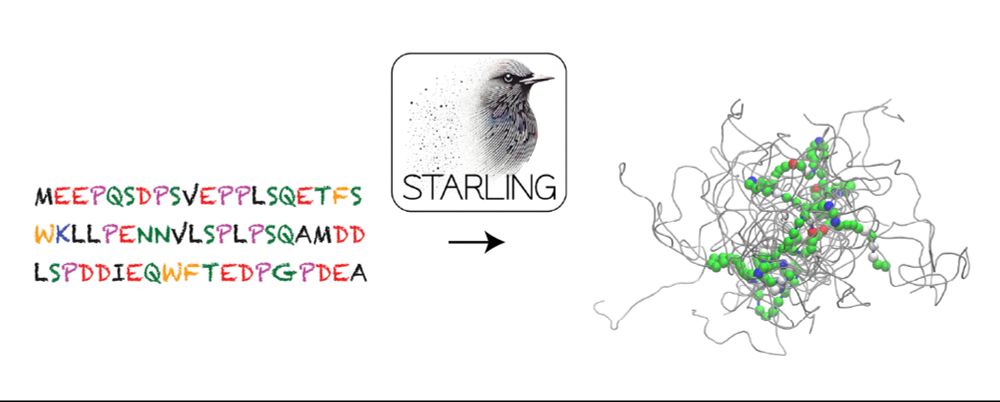
www.biorxiv.org/content/10.1...
In new work led by @liambai.bsky.social and me, we explore how sparse autoencoders can help us understand biology—going from mechanistic interpretability to mechanistic biology.
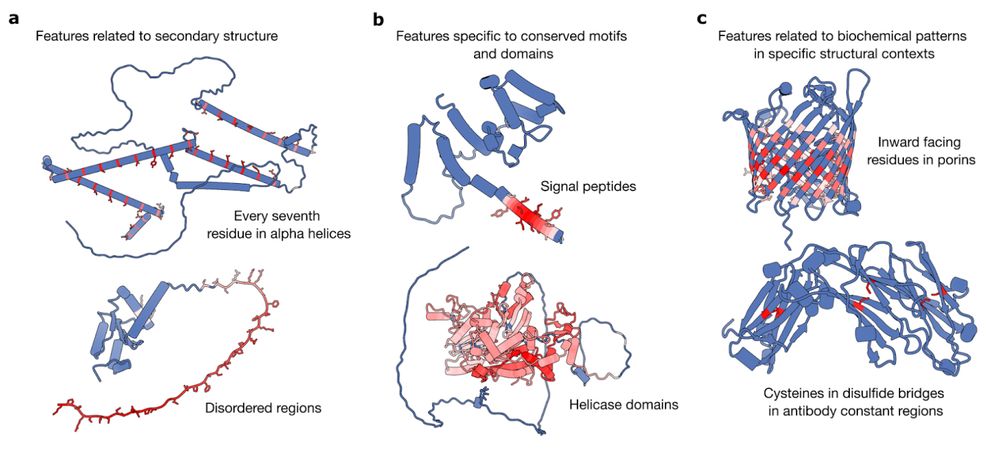
In new work led by @liambai.bsky.social and me, we explore how sparse autoencoders can help us understand biology—going from mechanistic interpretability to mechanistic biology.
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...