@embl.org @borklab.bsky.social
youtube.com/shorts/ymqyM...

#RExPO25 highlights with Markus List (@itisalist.bsky.social), Group Leader at @daisybio.de & Associate Professor at TUM.
Watch the full video for more insights ⤵️
youtu.be/iz7PisP_n08?...
#RExPO25 highlights with Markus List (@itisalist.bsky.social), Group Leader at @daisybio.de & Associate Professor at TUM.
Watch the full video for more insights ⤵️
youtu.be/iz7PisP_n08?...
Started as my Bachelor’s thesis — now it’s online:
doi.org/10.64898/202...
Huge thanks to @itisalist.bsky.social, @daisybio.de, @huckelhovenr.bsky.social , Felix Hoheneder & Johannes Kersting for all their help and support!

Started as my Bachelor’s thesis — now it’s online:
doi.org/10.64898/202...
Huge thanks to @itisalist.bsky.social, @daisybio.de, @huckelhovenr.bsky.social , Felix Hoheneder & Johannes Kersting for all their help and support!






great networking, beautiful location. Thanks to the organizers at @sib.swiss I appreciated especially today's session on startups in bioinformatics, that was insightful.

great networking, beautiful location. Thanks to the organizers at @sib.swiss I appreciated especially today's session on startups in bioinformatics, that was insightful.

🟣 @itisalist.bsky.social & Lisa Spindler (@daisybio.de)
🟣 Jan Baumbach & Fernando Delgado Chavez (@cosybio-uhh.bsky.social)
Check the full conference agenda ⤵️
repo4.eu/rexpo25/agen...
🇪🇺 #EUfunded #DrugRepurposing

🟣 @itisalist.bsky.social & Lisa Spindler (@daisybio.de)
🟣 Jan Baumbach & Fernando Delgado Chavez (@cosybio-uhh.bsky.social)
Check the full conference agenda ⤵️
repo4.eu/rexpo25/agen...
🇪🇺 #EUfunded #DrugRepurposing
academic.oup.com/bib/article/...
Bottom line: Use pseudobulk + DESeq2 in simple and pseudobulk + DREAM in more complex settings.
Collab w/ @leonhafner.bsky.social @itisalist.bsky.social

academic.oup.com/bib/article/...
Bottom line: Use pseudobulk + DESeq2 in simple and pseudobulk + DREAM in more complex settings.
Collab w/ @leonhafner.bsky.social @itisalist.bsky.social

🧵1/10


🔗 Attend at ISCB Nucleus: iscb.junolive.co
📍 If you’re not an ISCB member, register for access to ISCB Nucleus: lnkd.in/gMhrKGJz

🔗 Attend at ISCB Nucleus: iscb.junolive.co
📍 If you’re not an ISCB member, register for access to ISCB Nucleus: lnkd.in/gMhrKGJz

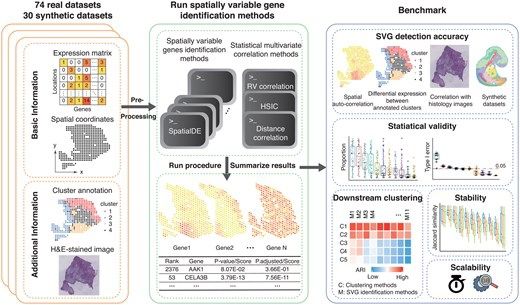




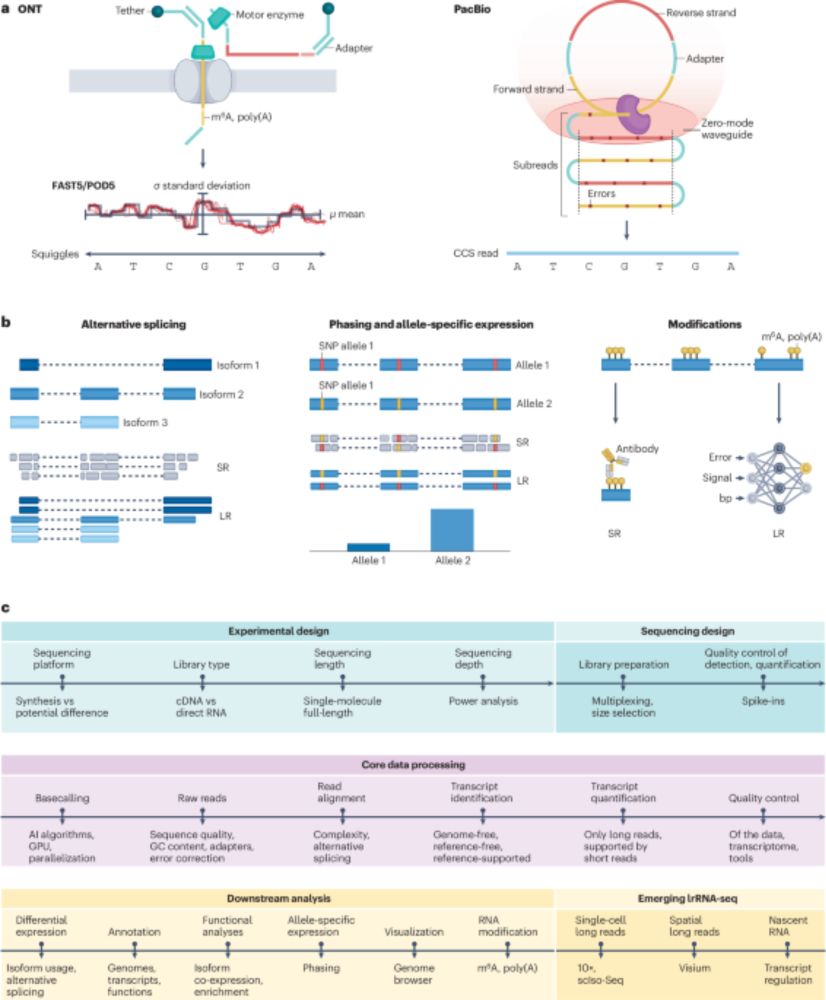

The start is done by @itisalist.bsky.social who heads the group. Markus joined TUM in 2018 and became a W2 tenure track associate professor in 2023. More members are about to follow! 📍

The start is done by @itisalist.bsky.social who heads the group. Markus joined TUM in 2018 and became a W2 tenure track associate professor in 2023. More members are about to follow! 📍