Topics include Kleb diversity, lineages, AMR, hypervirulence, how to use Kaptive & Kleborate for typing, and more!
klebnet.org/2025/11/18/k...
Topics include Kleb diversity, lineages, AMR, hypervirulence, how to use Kaptive & Kleborate for typing, and more!
klebnet.org/2025/11/18/k...

Norway’s strict antibiotic policies pay off. Record-low use in humans & animals, and some of Europe’s lowest #AMR rates. But ESBL & VRE are on the rise, so vigilance is vital.
www.unn.no/4a675d/sitea...
Norway’s strict antibiotic policies pay off. Record-low use in humans & animals, and some of Europe’s lowest #AMR rates. But ESBL & VRE are on the rise, so vigilance is vital.
www.unn.no/4a675d/sitea...
www.biorxiv.org/content/10.1...
github.com/iqbal-lab-or...

www.biorxiv.org/content/10.1...
github.com/iqbal-lab-or...
Ranging from methods development, microbiome, metagenomics and species specific (TB, Strep pneumo, measles) projects

Ranging from methods development, microbiome, metagenomics and species specific (TB, Strep pneumo, measles) projects
We hope this will be useful
- short reads, long reads, hybrid assembly or all combined - for your own tools/validations 🥳 #Microsky
journals.asm.org/doi/10.1128/...

We hope this will be useful
- short reads, long reads, hybrid assembly or all combined - for your own tools/validations 🥳 #Microsky
journals.asm.org/doi/10.1128/...

interpretamr.github.io/AMRrules

interpretamr.github.io/AMRrules
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
@irenlohr.bsky.social @katholt.bsky.social
genomemedicine.biomedcentral.com/articles/10....
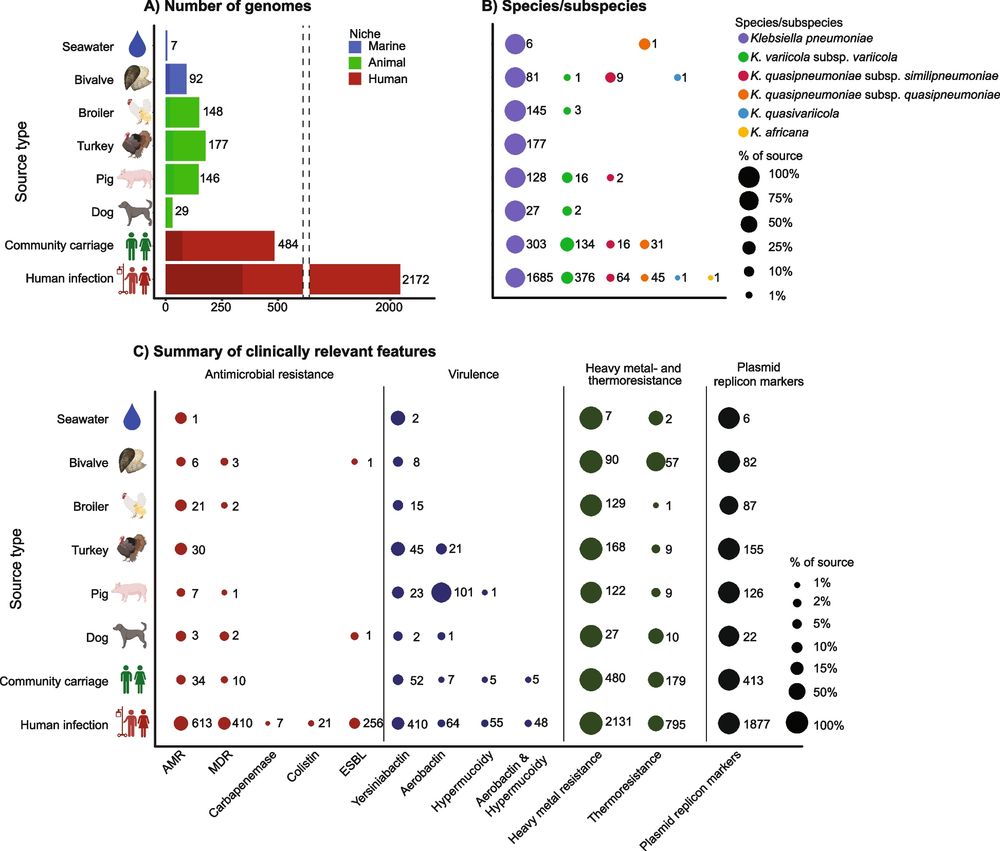
@irenlohr.bsky.social @katholt.bsky.social
genomemedicine.biomedcentral.com/articles/10....
Big thanks to coauthors @kelwyres.bsky.social, @katholt.bsky.social, @genomarit.bsky.social and Iren Löhr.
Here's what we did to improve in silico antigen typing 👇🧵
www.biorxiv.org/content/10.1...

Big thanks to coauthors @kelwyres.bsky.social, @katholt.bsky.social, @genomarit.bsky.social and Iren Löhr.
Here's what we did to improve in silico antigen typing 👇🧵
www.biorxiv.org/content/10.1...
I'm excited to announce Autocycler, my new tool for consensus assembly of long-read bacterial genomes!
It's the successor to Trycycler, designed to be faster and less reliant on user intervention.
Check it out: github.com/rrwick/Autoc...
(1/5)
I'm excited to announce Autocycler, my new tool for consensus assembly of long-read bacterial genomes!
It's the successor to Trycycler, designed to be faster and less reliant on user intervention.
Check it out: github.com/rrwick/Autoc...
(1/5)
www.regjeringen.no/en/dokumente...

www.regjeringen.no/en/dokumente...
2009 was a big year for Klebs!
- First genome published
- First description of ST258 KPC-producing clone
pubmed.ncbi.nlm.nih.gov/19218573/
- First description of NDM-1 beta-lactamase
pubmed.ncbi.nlm.nih.gov/19770275/
2009 was a big year for Klebs!
- First genome published
- First description of ST258 KPC-producing clone
pubmed.ncbi.nlm.nih.gov/19218573/
- First description of NDM-1 beta-lactamase
pubmed.ncbi.nlm.nih.gov/19770275/
www.biorxiv.org/content/10.1...
Presented by Marit Hetland #Klebsiella2024

www.biorxiv.org/content/10.1...
Presented by Marit Hetland #Klebsiella2024

#microsky
www.findaphd.com/phds/project...
#microsky
www.findaphd.com/phds/project...

