Gaurav Bhardwaj
@gauravbhardwaj.bsky.social
Assistant Professor of Medicinal Chemistry at Univ of Washington and Institute for Protein Design | Scientist | Computational Peptide/Protein Design
Glad that we could contribute to this very cool work by @yehlincho.bsky.social and @sokrypton.org! See the nice skeetorial below by Yehlin. More to come soon with experimental validation and exciting applications in protein and peptide design.
Thrilled to announce our new preprint, “Protein Hunter: Exploiting Structure Hallucination within Diffusion for Protein Design,” in collaboration with @Griffin, @GBhardwaj8 and @sokrypton.org
🧬Code and notebooks will be released by the end of this week.
🎧Golden- Kpop Demon Hunters
🧬Code and notebooks will be released by the end of this week.
🎧Golden- Kpop Demon Hunters
October 13, 2025 at 5:53 PM
Glad that we could contribute to this very cool work by @yehlincho.bsky.social and @sokrypton.org! See the nice skeetorial below by Yehlin. More to come soon with experimental validation and exciting applications in protein and peptide design.
Reposted by Gaurav Bhardwaj
(1/7)
Training biomolecular foundation models shouldn't be so hard. And open-source structure prediction is important. So today we're releasing two software packages: AtomWorks and RosettaFold3 (RF3)
[https://www.biorxiv.org/content/10.1101/2025.08.14.670328v2](www.biorxiv.org/content/10.1...)
Training biomolecular foundation models shouldn't be so hard. And open-source structure prediction is important. So today we're releasing two software packages: AtomWorks and RosettaFold3 (RF3)
[https://www.biorxiv.org/content/10.1101/2025.08.14.670328v2](www.biorxiv.org/content/10.1...)

Accelerating Biomolecular Modeling with AtomWorks and RF3
Deep learning methods trained on protein structure databases have revolutionized biomolecular structure prediction, but developing and training new models remains a considerable challenge. To facilita...
www.biorxiv.org
August 15, 2025 at 5:17 PM
(1/7)
Training biomolecular foundation models shouldn't be so hard. And open-source structure prediction is important. So today we're releasing two software packages: AtomWorks and RosettaFold3 (RF3)
[https://www.biorxiv.org/content/10.1101/2025.08.14.670328v2](www.biorxiv.org/content/10.1...)
Training biomolecular foundation models shouldn't be so hard. And open-source structure prediction is important. So today we're releasing two software packages: AtomWorks and RosettaFold3 (RF3)
[https://www.biorxiv.org/content/10.1101/2025.08.14.670328v2](www.biorxiv.org/content/10.1...)
Reposted by Gaurav Bhardwaj
We are very excited to announce that early bird registration for European RosettaCon 2025 is now open!
More information here: europeanrosettacon.org
More information here: europeanrosettacon.org
June 30, 2025 at 1:56 PM
We are very excited to announce that early bird registration for European RosettaCon 2025 is now open!
More information here: europeanrosettacon.org
More information here: europeanrosettacon.org
Great to have our manuscript with @sokrypton.org 's lab describing AfCyDesign finally out in @natcomms.bsky.social . Structure prediction, sequence redesign, de novo hallucination of cyclic peptides, and some binder design examples in this version.
rdcu.be/em0vA
rdcu.be/em0vA
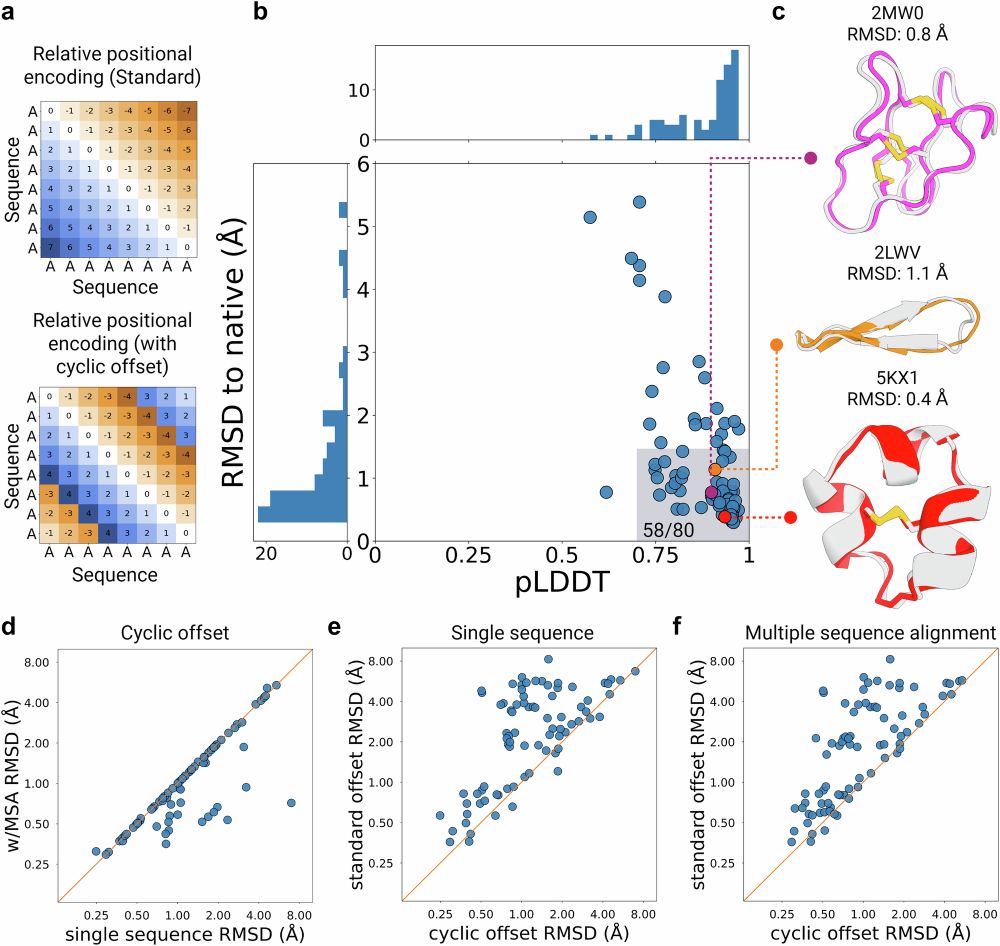
Cyclic peptide structure prediction and design using AlphaFold2
Nature Communications - AfCycDesign: Cyclic offset to the relative positional encoding in AlphaFold2 enables accurate structure prediction, sequence redesign, and de novo hallucination of cyclic...
rdcu.be
May 23, 2025 at 7:07 PM
Great to have our manuscript with @sokrypton.org 's lab describing AfCyDesign finally out in @natcomms.bsky.social . Structure prediction, sequence redesign, de novo hallucination of cyclic peptides, and some binder design examples in this version.
rdcu.be/em0vA
rdcu.be/em0vA
Reposted by Gaurav Bhardwaj
1/ In two back-to-back papers, we present our de novo TRACeR platform for targeting MHC-I and MHC-II antigens
TRACeR for MHC-I: go.nature.com/4gcLzn5
TRACeR for MHC-II: go.nature.com/4gj5OQk
TRACeR for MHC-I: go.nature.com/4gcLzn5
TRACeR for MHC-II: go.nature.com/4gj5OQk
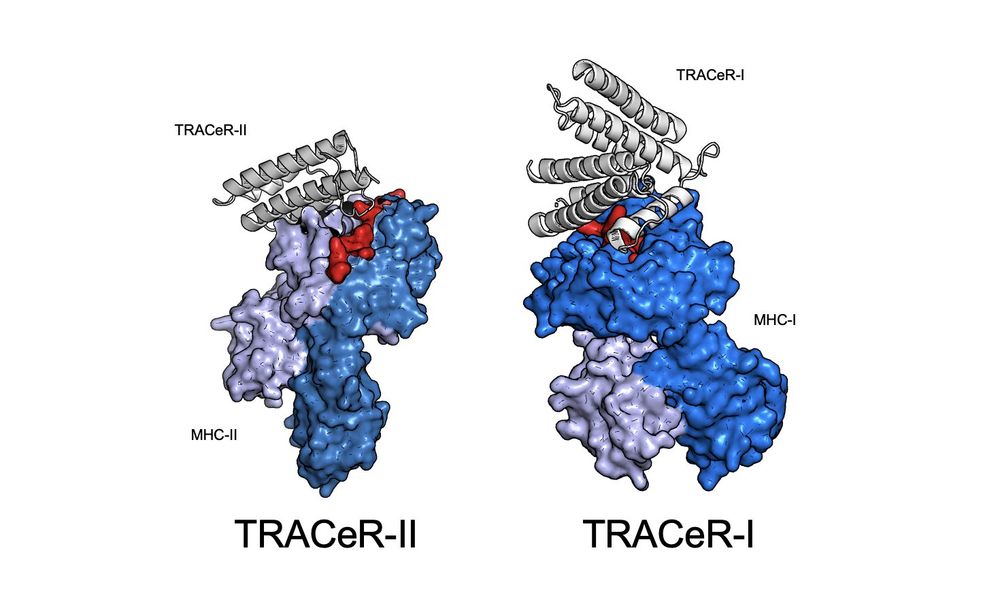
December 17, 2024 at 12:56 AM
1/ In two back-to-back papers, we present our de novo TRACeR platform for targeting MHC-I and MHC-II antigens
TRACeR for MHC-I: go.nature.com/4gcLzn5
TRACeR for MHC-II: go.nature.com/4gj5OQk
TRACeR for MHC-I: go.nature.com/4gcLzn5
TRACeR for MHC-II: go.nature.com/4gj5OQk
Reposted by Gaurav Bhardwaj
Today, several #NobelPrize Laureates arrive in Stockholm, warmly welcomed by Hans Ellegren. Here we see David Baker stepping off the plane at Arlanda.
This week is packed with inspiration, press conferences and lectures, so stay tuned! 🌟
@uofwa.bsky.social @hhmi.bsky.social
#Science #AcademicSky
This week is packed with inspiration, press conferences and lectures, so stay tuned! 🌟
@uofwa.bsky.social @hhmi.bsky.social
#Science #AcademicSky

December 5, 2024 at 12:48 PM
Today, several #NobelPrize Laureates arrive in Stockholm, warmly welcomed by Hans Ellegren. Here we see David Baker stepping off the plane at Arlanda.
This week is packed with inspiration, press conferences and lectures, so stay tuned! 🌟
@uofwa.bsky.social @hhmi.bsky.social
#Science #AcademicSky
This week is packed with inspiration, press conferences and lectures, so stay tuned! 🌟
@uofwa.bsky.social @hhmi.bsky.social
#Science #AcademicSky
Hats off to the people compiling all those starter packs—it has made the move to this site so much easier!
November 21, 2024 at 5:52 PM
Hats off to the people compiling all those starter packs—it has made the move to this site so much easier!
Here goes the skeetorial for the latest preprint from our lab describing RFpeptides, a pipeline for design of target-binding macrocycles using diffusion models. Big shoutout to Stephen Rettie, David Juergens, Victor Adebomi for leading the project (1/n)
Preprint link: www.biorxiv.org/content/10.1...
Preprint link: www.biorxiv.org/content/10.1...
www.biorxiv.org
November 21, 2024 at 6:21 AM
Here goes the skeetorial for the latest preprint from our lab describing RFpeptides, a pipeline for design of target-binding macrocycles using diffusion models. Big shoutout to Stephen Rettie, David Juergens, Victor Adebomi for leading the project (1/n)
Preprint link: www.biorxiv.org/content/10.1...
Preprint link: www.biorxiv.org/content/10.1...
Super excited to share the latest preprint from our lab on macrocycle binder design! Skeetorial (or whatever they are called) to follow soon. Thanks to all the amazing collaborators!
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

Accurate de novo design of high-affinity protein binding macrocycles using deep learning
The development of macrocyclic binders to therapeutic proteins has typically relied on large-scale screening methods that are resource-intensive and provide little control over binding mode. Despite c...
www.biorxiv.org
November 19, 2024 at 7:09 PM
Super excited to share the latest preprint from our lab on macrocycle binder design! Skeetorial (or whatever they are called) to follow soon. Thanks to all the amazing collaborators!
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

