Gaurav Bhardwaj
@gauravbhardwaj.bsky.social
Assistant Professor of Medicinal Chemistry at Univ of Washington and Institute for Protein Design | Scientist | Computational Peptide/Protein Design
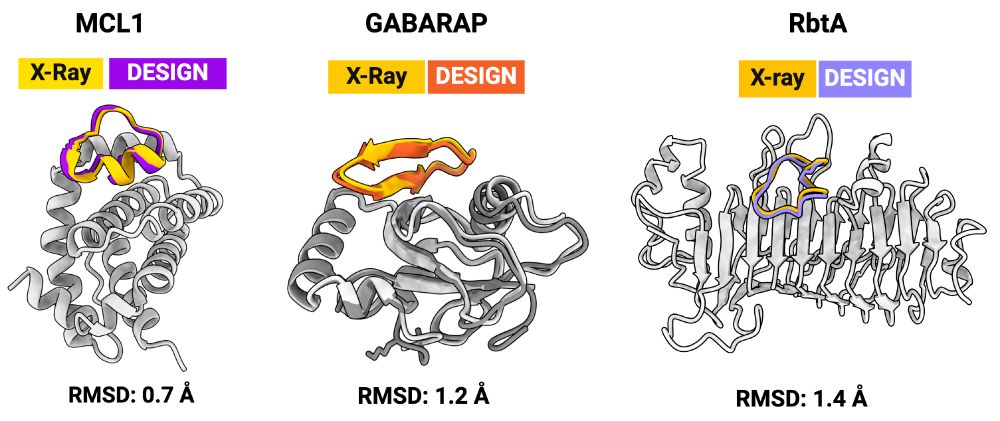
X-ray structures for the macrocycle bound complexes also match very closely with the design models (CA RMSD < 1.5 angstroms). The designs are diverse: helix-containing (MCL-1/Mdm2), beta-strands (GABARAP), and loopy (RbtA).
November 21, 2024 at 6:21 AM
X-ray structures for the macrocycle bound complexes also match very closely with the design models (CA RMSD < 1.5 angstroms). The designs are diverse: helix-containing (MCL-1/Mdm2), beta-strands (GABARAP), and loopy (RbtA).
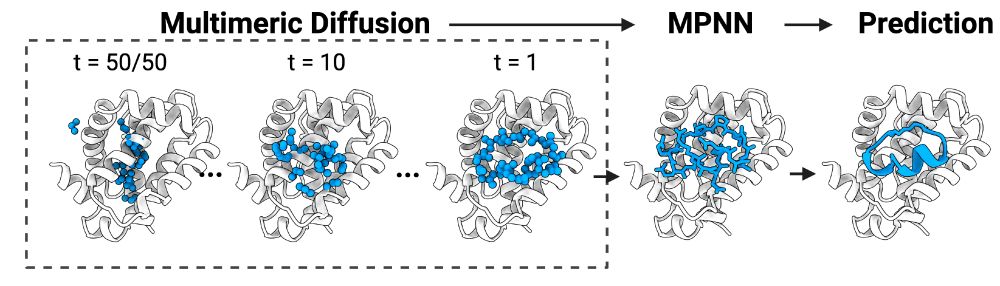
Here, we modified RFdiffusion positional encodings to design cyclic peptide backbones against selected targets, followed by sequence design using ProteinMPNN. Final designs were selected based of confidence metrics from AF2/RF2 re-prediction and Rosetta-based interface quality metrics.
November 21, 2024 at 6:21 AM
Here, we modified RFdiffusion positional encodings to design cyclic peptide backbones against selected targets, followed by sequence design using ProteinMPNN. Final designs were selected based of confidence metrics from AF2/RF2 re-prediction and Rosetta-based interface quality metrics.
Here is a GIF in the meantime:
November 19, 2024 at 7:09 PM
Here is a GIF in the meantime:

