Gaurav Bhardwaj
@gauravbhardwaj.bsky.social
Assistant Professor of Medicinal Chemistry at Univ of Washington and Institute for Protein Design | Scientist | Computational Peptide/Protein Design
Thank you!! Look forward to meeting you in Grenoble soon.
May 27, 2025 at 1:38 AM
Thank you!! Look forward to meeting you in Grenoble soon.
Thanks for the shoutout! I agree - We have not tried it, but
don't expect it to work well for disordered targets with low-throughput testing. Not yet, at least! 😀
don't expect it to work well for disordered targets with low-throughput testing. Not yet, at least! 😀
November 21, 2024 at 5:57 PM
Thanks for the shoutout! I agree - We have not tried it, but
don't expect it to work well for disordered targets with low-throughput testing. Not yet, at least! 😀
don't expect it to work well for disordered targets with low-throughput testing. Not yet, at least! 😀
Ha ha Probably not! :)
November 21, 2024 at 6:28 AM
Ha ha Probably not! :)
I may have figured out how to add a GIF of diffusion trajectory. Lets see! 😀
i.giphy.com/media/v1.Y2l...
i.giphy.com/media/v1.Y2l...
giphy.com
November 21, 2024 at 6:21 AM
I may have figured out how to add a GIF of diffusion trajectory. Lets see! 😀
i.giphy.com/media/v1.Y2l...
i.giphy.com/media/v1.Y2l...
It is a really fun time to be designing peptides/proteins. Please reach out if you have targets you would like to design macrocycle binders.
November 21, 2024 at 6:21 AM
It is a really fun time to be designing peptides/proteins. Please reach out if you have targets you would like to design macrocycle binders.
None of this would have been possible without all the great collaborators at @uwproteindesign.bsky.social and beyond (still trying to find everyone here!). There is much more to come as we continue to fine-tune and expand RFpeptides.
November 21, 2024 at 6:21 AM
None of this would have been possible without all the great collaborators at @uwproteindesign.bsky.social and beyond (still trying to find everyone here!). There is much more to come as we continue to fine-tune and expand RFpeptides.
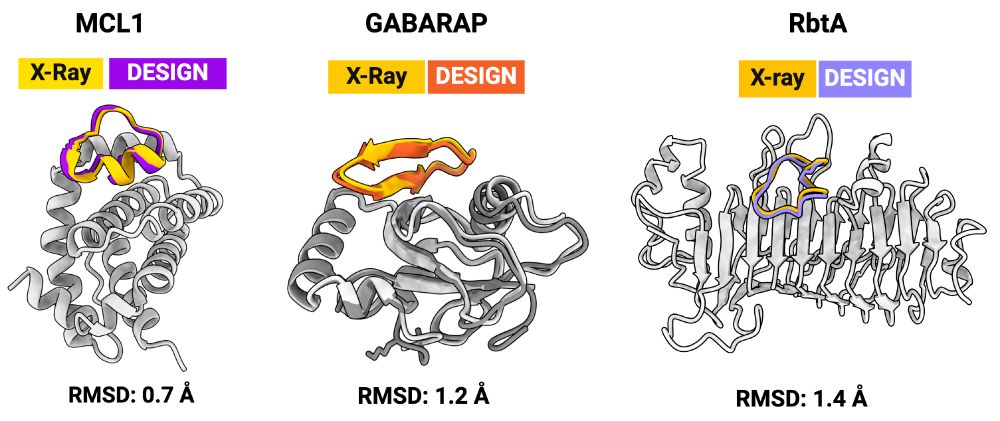
Perhaps the most fun part for us was the RbtA, where we did not have the target structure available when we designed against it. So we predicted the structure using AF2/RF2 and then designed against the predicted structures. Tested < 15 designs and got a Kd <10 nM binder!
November 21, 2024 at 6:21 AM
Perhaps the most fun part for us was the RbtA, where we did not have the target structure available when we designed against it. So we predicted the structure using AF2/RF2 and then designed against the predicted structures. Tested < 15 designs and got a Kd <10 nM binder!
X-ray structures for the macrocycle bound complexes also match very closely with the design models (CA RMSD < 1.5 angstroms). The designs are diverse: helix-containing (MCL-1/Mdm2), beta-strands (GABARAP), and loopy (RbtA).
November 21, 2024 at 6:21 AM
X-ray structures for the macrocycle bound complexes also match very closely with the design models (CA RMSD < 1.5 angstroms). The designs are diverse: helix-containing (MCL-1/Mdm2), beta-strands (GABARAP), and loopy (RbtA).
We used RFpeptides to design binders against four different targets: Mdm2, MCL-1, GABARAP, and RbtA. For each of the targets, we experimemtally tested <20 designs. We got 1-10 micromolar Kd binders against Mdm2 and MCL-1, and 1-10 nM binders against GABARAP and RbtA.
November 21, 2024 at 6:21 AM
We used RFpeptides to design binders against four different targets: Mdm2, MCL-1, GABARAP, and RbtA. For each of the targets, we experimemtally tested <20 designs. We got 1-10 micromolar Kd binders against Mdm2 and MCL-1, and 1-10 nM binders against GABARAP and RbtA.
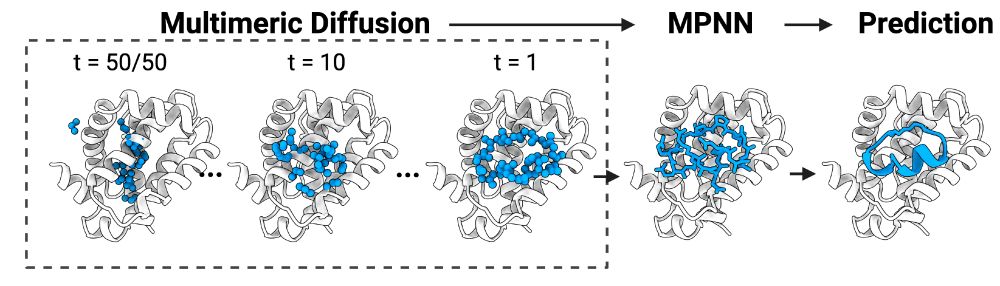
Here, we modified RFdiffusion positional encodings to design cyclic peptide backbones against selected targets, followed by sequence design using ProteinMPNN. Final designs were selected based of confidence metrics from AF2/RF2 re-prediction and Rosetta-based interface quality metrics.
November 21, 2024 at 6:21 AM
Here, we modified RFdiffusion positional encodings to design cyclic peptide backbones against selected targets, followed by sequence design using ProteinMPNN. Final designs were selected based of confidence metrics from AF2/RF2 re-prediction and Rosetta-based interface quality metrics.
So the design pipeline has to be good at designing and also selecting the best 10-20 binders. And it should also work for diverse targets. RFpeptides seems to be able to address a lot of those early issues and meet the requirements.
November 21, 2024 at 6:21 AM
So the design pipeline has to be good at designing and also selecting the best 10-20 binders. And it should also work for diverse targets. RFpeptides seems to be able to address a lot of those early issues and meet the requirements.
Why are we excited about it? Well, we spent a lot of effort over the years to accurately design high-affinity binders with our physics-based methods without much success. Since we rely on chemical synthesis of macrocycles, we were limited to making and testing only 10-20 designs/target in our lab.
November 21, 2024 at 6:21 AM
Why are we excited about it? Well, we spent a lot of effort over the years to accurately design high-affinity binders with our physics-based methods without much success. Since we rely on chemical synthesis of macrocycles, we were limited to making and testing only 10-20 designs/target in our lab.
Here is a GIF in the meantime:
November 19, 2024 at 7:09 PM
Here is a GIF in the meantime:

