academic.oup.com/mbe/advance-...

academic.oup.com/mbe/advance-...
genomebiology.biomedcentral.com/articles/10....

genomebiology.biomedcentral.com/articles/10....

www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
arxiv.org/abs/2506.12986

arxiv.org/abs/2506.12986

Its thought deep learning will substantially improve PGS but the reality is MANY have tried but no/little gain has been seen so far. Here we report our negative results.
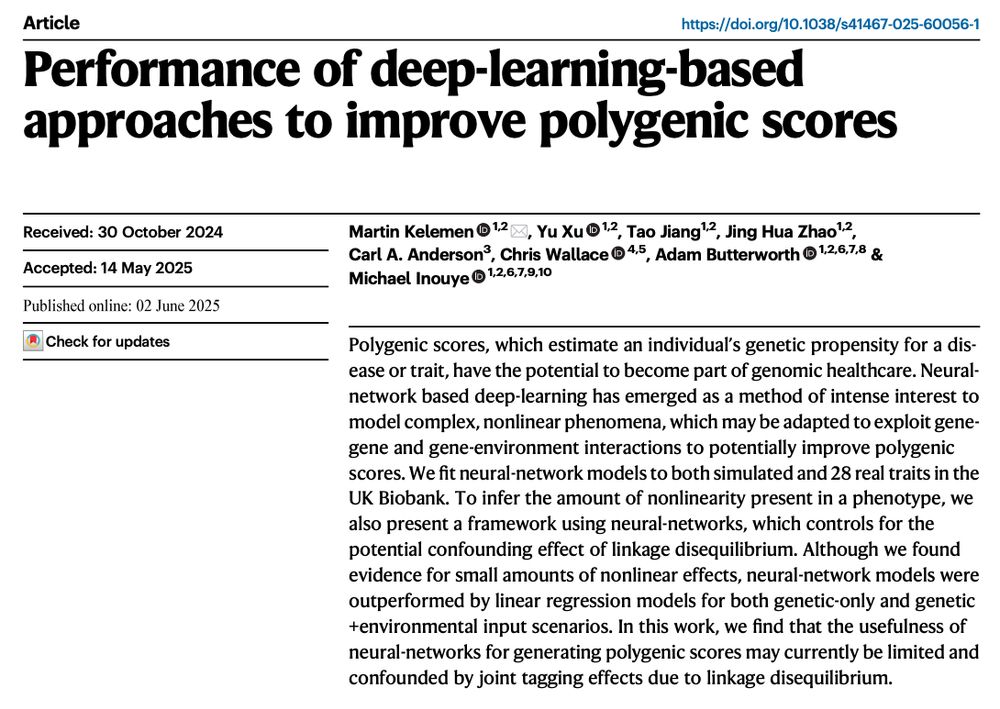
Its thought deep learning will substantially improve PGS but the reality is MANY have tried but no/little gain has been seen so far. Here we report our negative results.
longcallR: github.com/huangnengCSU...
Nextflow workflow: github.com/huangnengCSU...


longcallR: github.com/huangnengCSU...
Nextflow workflow: github.com/huangnengCSU...
📖 Read the blog post: hubs.la/Q03psL-t0

📖 Read the blog post: hubs.la/Q03psL-t0
Here, we used simulation work + longitudinal GWAS and downstream analyses to explore risks involved in cognitive/physical decline
(1/)🧵🧵
shorturl.at/99gqL

Here, we used simulation work + longitudinal GWAS and downstream analyses to explore risks involved in cognitive/physical decline
(1/)🧵🧵
shorturl.at/99gqL

Call for abstracts and registrations coming soon!

Call for abstracts and registrations coming soon!
Tldr: Accurate ploidy is important for reducing false positives; appropriate alignment reduces false negatives.
www.biorxiv.org/content/10.1...

Tldr: Accurate ploidy is important for reducing false positives; appropriate alignment reduces false negatives.
www.biorxiv.org/content/10.1...





www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
🔍 Workflow view – Visualize logical structure of pipelines
⚙️ Process view – See all processes in one place
🌐 Seqera view – Manage connected CEs directly in the IDE
📚 Resources view – Quickly access Copilot, AI tools & training
🔍 Workflow view – Visualize logical structure of pipelines
⚙️ Process view – See all processes in one place
🌐 Seqera view – Manage connected CEs directly in the IDE
📚 Resources view – Quickly access Copilot, AI tools & training

We lay out best-practice guidelines for releasing variant effect predictors, developed through the Atlas of Variant Effects Alliance @varianteffect.bsky.social
Open, interpretable, and clinically useful VEPs are the goal.
📄 doi.org/10.1186/s130...

We lay out best-practice guidelines for releasing variant effect predictors, developed through the Atlas of Variant Effects Alliance @varianteffect.bsky.social
Open, interpretable, and clinically useful VEPs are the goal.
📄 doi.org/10.1186/s130...
@ewels.bsky.social and Ben Sherman discuss the upcoming #Nextflow strict syntax - cleaner code, better errors, and a more consistent language framework.
Learn what's changing and how to prepare your pipelines: hubs.la/Q03hpNvx0

@ewels.bsky.social and Ben Sherman discuss the upcoming #Nextflow strict syntax - cleaner code, better errors, and a more consistent language framework.
Learn what's changing and how to prepare your pipelines: hubs.la/Q03hpNvx0