Romix Bio | Tech Inno @ NYGC | PhD @ Cornell BME | postbac @ NHGRI
mckellardw.github.io
So far, across the ~120 registrants we have 7 academic institutions, 4 companies, and 2 journals represented!
Talks from Chris Mason, Ildar Gainetdinov,
@xuebingwu.bsky.social, and Sanja Vickovic!
Come down to @nygenome.org and hear about the latest in RNA technology development being done in NYC!

So far, across the ~120 registrants we have 7 academic institutions, 4 companies, and 2 journals represented!

Talks from Chris Mason, Ildar Gainetdinov,
@xuebingwu.bsky.social, and Sanja Vickovic!
Come down to @nygenome.org and hear about the latest in RNA technology development being done in NYC!

Talks from Chris Mason, Ildar Gainetdinov,
@xuebingwu.bsky.social, and Sanja Vickovic!
Come down to @nygenome.org and hear about the latest in RNA technology development being done in NYC!
Imagination and new opportunities go ⏫
@landau.bsky.social
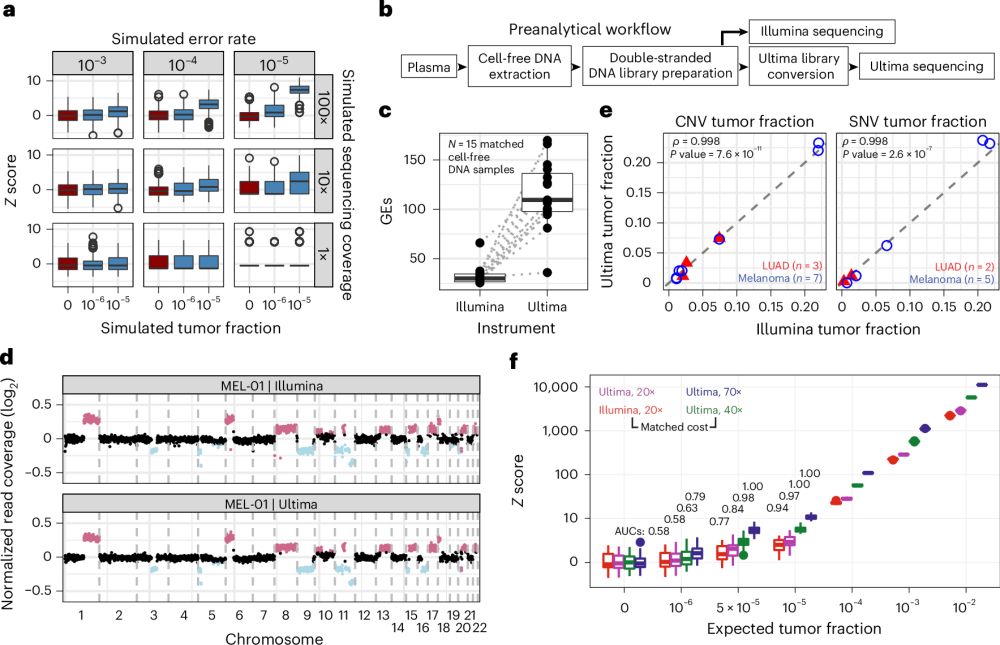
Let's look at what you can do when you perform deep WGS on cfDNA for cancer detection. Our recent work with the
@UltimaGenomics
platform 🧵👇
Lets goooo!!
rdcu.be/ehkUb
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...


MultiPerturb-seq 🎛️ ❌ ↕️
Over the last few years, we've been combining CRISPR screens with multimodal readouts of gene expression (RNA) and chromatin accessibility (DNA). In this study, we bring those together within the same cells.
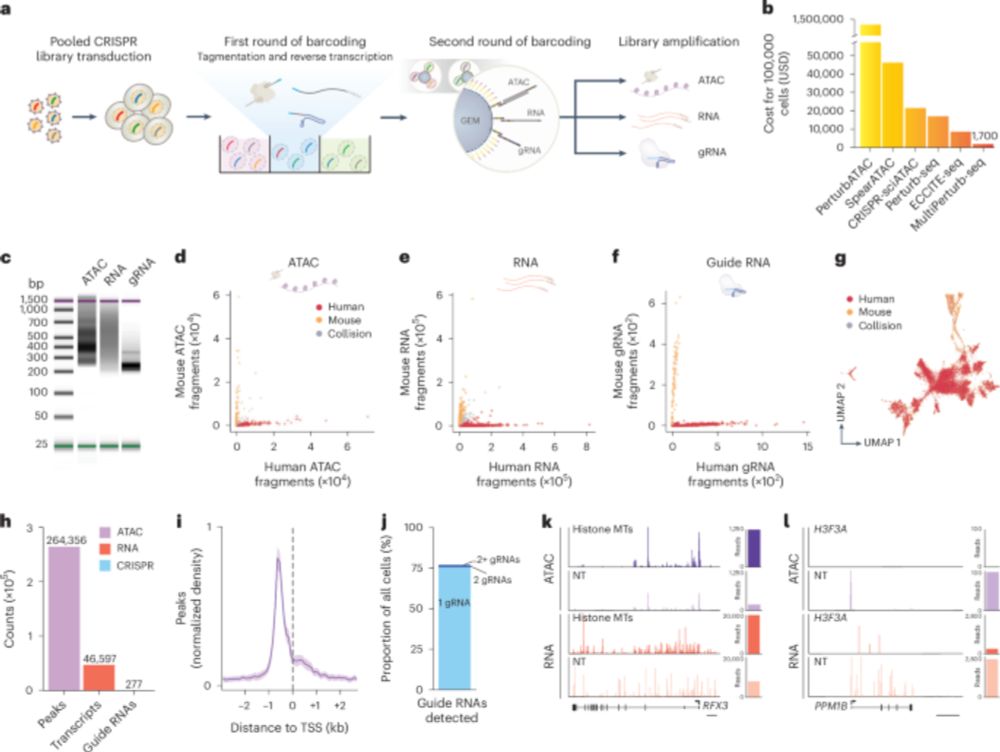
MultiPerturb-seq 🎛️ ❌ ↕️
Over the last few years, we've been combining CRISPR screens with multimodal readouts of gene expression (RNA) and chromatin accessibility (DNA). In this study, we bring those together within the same cells.
#RNAsky #RNAbiology

Our new method to map the host-microbiome interface at high-resolution, now on bioRxiv!
Co-led with Lena Takayasu & @dwmckellar.bsky.social collab between the De Vlaminck and Brito labs
doi.org/10.1101/2024...
A thread⬇️

Our new method to map the host-microbiome interface at high-resolution, now on bioRxiv!
Co-led with Lena Takayasu & @dwmckellar.bsky.social collab between the De Vlaminck and Brito labs
doi.org/10.1101/2024...
A thread⬇️

