Previously PhD @imperialcollegeldn.bsky.social.
Modelling gene regulation in heterogeneous cell populations.


go.nature.com/4lFmpiN

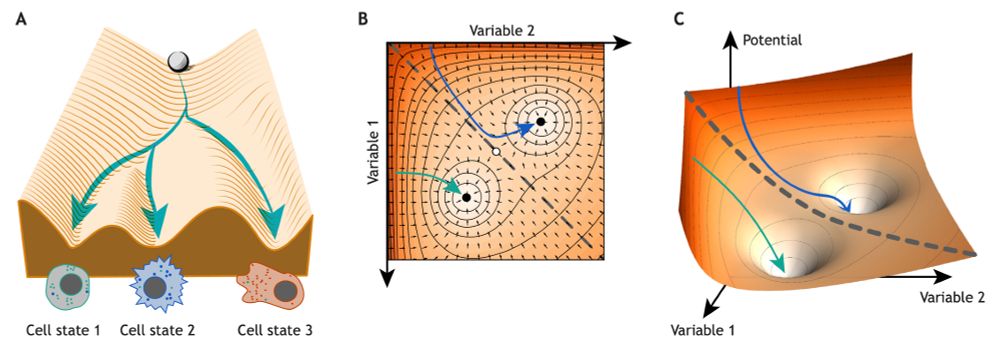
Call to action from the community to achive this ambitious goal!
Call to action from the community to achive this ambitious goal!
time.com/7283507/safe...

time.com/7283507/safe...
Quite an indictment of some of the current single cell "virtual cell" foundation models. Even for the relatively mundane applications, cell labeling, batch correction etc, they are poor compared to much simpler & cheaper methods.
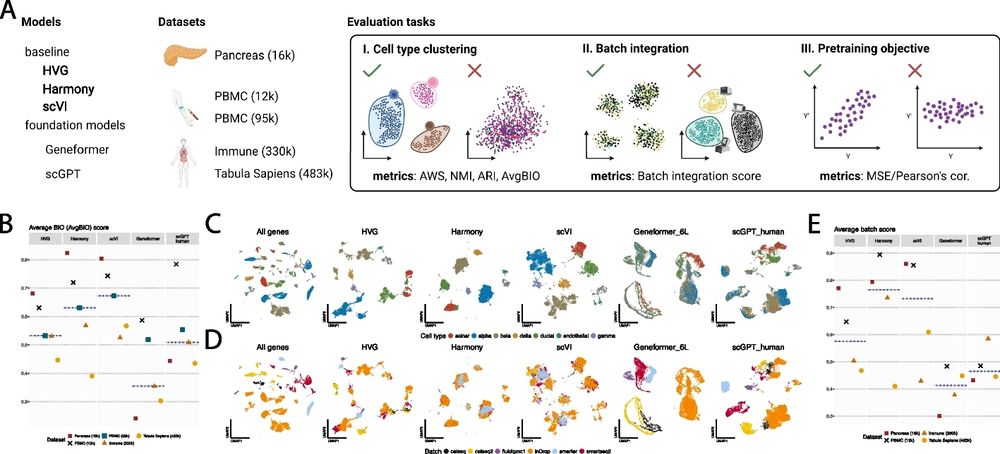
Quite an indictment of some of the current single cell "virtual cell" foundation models. Even for the relatively mundane applications, cell labeling, batch correction etc, they are poor compared to much simpler & cheaper methods.
Read this @reviewcommons.org transfer Research Article by @mar-ferrando-marco.bsky.social & Michalis Barkoulas @imperialcollegeldn.bsky.social
doi.org/10.1242/dev....
![figure 3 (C) Representative images of LIT-1::GFP following the L2 asymmetric division in controls (WT), tissue-specific efl-3 mutants [efl-3(−)] and animals containing the lit-1(icb164) CRISPR deletion. Seam cell nuclei are circled in white. Hypodermal nuclei are labelled with blue arrowheads. a, anterior daughter cell; p, posterior daughter cell.](https://cdn.bsky.app/img/feed_thumbnail/plain/did:plc:btitwyhiegkb6f62oguxt6m6/bafkreih2xjdz4bhvm7z7h5d7i3gx4qw2xrfhix3iphhvaddvhaogimnz4y@jpeg)
Read this @reviewcommons.org transfer Research Article by @mar-ferrando-marco.bsky.social & Michalis Barkoulas @imperialcollegeldn.bsky.social
doi.org/10.1242/dev....
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...

