Well, I discovered there is a name for this pattern: Ralph. We made a Ralph Wiggum deep researcher. monarch-initiative.github.io/deep-researc...

Well, I discovered there is a name for this pattern: Ralph. We made a Ralph Wiggum deep researcher. monarch-initiative.github.io/deep-researc...
A solid machine learning framework & to predict strain-level phage-host interactions across diverse bacterial genera from genome sequences alone. Avery Noonan from the Arkin Lab led this massive effort
www.biorxiv.org/content/10.1...
A solid machine learning framework & to predict strain-level phage-host interactions across diverse bacterial genera from genome sequences alone. Avery Noonan from the Arkin Lab led this massive effort
www.biorxiv.org/content/10.1...

Our new paper by Mannion et al. takes a systematic look at "hidden enhancers" and why they remain so hard to find. With @mosterwalder.bsky.social, @jlopezrios.bsky.social & many more
www.nature.com/articles/s41...

Our new paper by Mannion et al. takes a systematic look at "hidden enhancers" and why they remain so hard to find. With @mosterwalder.bsky.social, @jlopezrios.bsky.social & many more
www.nature.com/articles/s41...

By @claudia-lopez.bsky.social
www.thetransmitter.org/community/ha...

knowledgegraphinsights.com/chris-mungall/

knowledgegraphinsights.com/chris-mungall/
Ruth has contributed extensively to the curation of key resources such as HGNC, Gene Ontology (GO), and IMEx, and has been instrumental in developing curation standards. Ruth is a past chair of the ISB EC.

Ruth has contributed extensively to the curation of key resources such as HGNC, Gene Ontology (GO), and IMEx, and has been instrumental in developing curation standards. Ruth is a past chair of the ISB EC.
Kimberly's career reflects expertise, sustained innovation, & dedicated service to community. She's contributed to many projects, including WormBase, the Gene Ontology, & the Alliance of Genome Resources.

Kimberly's career reflects expertise, sustained innovation, & dedicated service to community. She's contributed to many projects, including WormBase, the Gene Ontology, & the Alliance of Genome Resources.
Tiago's a passionate and motivated scientist interested in linked open data, ontologies, the semantic web, and their application in modeling cells and cell types. He is active in many curation projects & with ISB.

Tiago's a passionate and motivated scientist interested in linked open data, ontologies, the semantic web, and their application in modeling cells and cell types. He is active in many curation projects & with ISB.
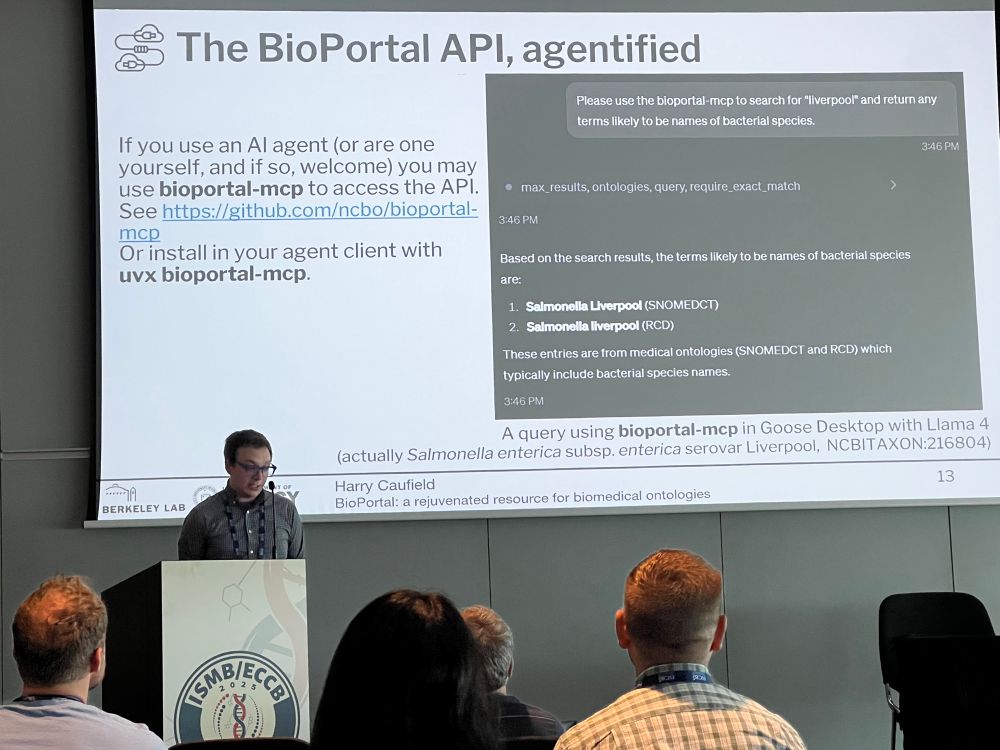


More info: www.open-bio.org/events/bosc-...
(2/2)

If this is how they treat Alex Padilla, a United States Senator, how do you think they’ll treat you?

www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...

That's when you evade the rules of one place or territory by outsourcing your needs or dirty deeds to another territory.
El Salvador has become a Network State for Concentration Camps. Expect more moves like this.
I'm flying to El Salvador tomorrow morning to check on his condition and discuss his return.
That's when you evade the rules of one place or territory by outsourcing your needs or dirty deeds to another territory.
El Salvador has become a Network State for Concentration Camps. Expect more moves like this.

