


![code segment from collab notebook:
output = dna_model.predict_sequence(
sequence='GATTACA'.center(2048, 'N'), # Pad to valid sequence length.
requested_outputs=[dna_client.OutputType.DNASE],
ontology_terms=['UBERON:0002048'], # Lung.
)](https://cdn.bsky.app/img/feed_thumbnail/plain/did:plc:dmaknbliyv4xeomex3zwa4gq/bafkreidomwlro7gnyfwblhkdd6ry3tadxi7u36brjoksoibrnl4xhjxjza@jpeg)





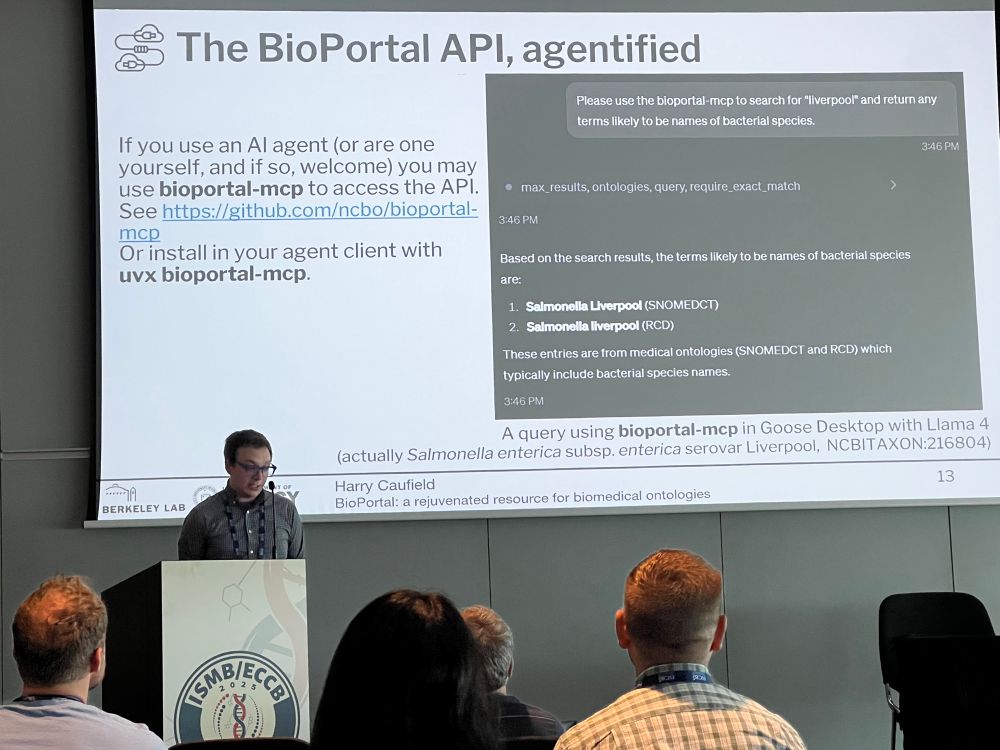
academic.oup.com/bioinformati.... Great work from Harry Caufield who led the study, and all the authors. SPIRES uses a schema and ontology driven approach to extract complex knowledge nuggets from text.

academic.oup.com/bioinformati.... Great work from Harry Caufield who led the study, and all the authors. SPIRES uses a schema and ontology driven approach to extract complex knowledge nuggets from text.

















