
Postdoc at Big Data Biology Lab

@bigdatabiology.bsky.social lab in Shanghai see the light! The work includes my three favorite things research-wise: 🦠 microbiome, 🧬 long-reads, and 🐶 dogs.
See our new preprint:
www.biorxiv.org/content/10.1...
• Shanghai dog microbiome / @annacusco.bsky.social
• microbetag / @hariszaf.bsky.social
• invitation for the #MVIF 42 / @microbiomevif.bsky.social
& more.

• Shanghai dog microbiome / @annacusco.bsky.social
• microbetag / @hariszaf.bsky.social
• invitation for the #MVIF 42 / @microbiomevif.bsky.social
& more.
I am currently looking for my next research adventure. If you have insights on the microbiome/microbial genomics job market in Europe (academia & industry), or want to chat about this work, please reach out.


I am currently looking for my next research adventure. If you have insights on the microbiome/microbial genomics job market in Europe (academia & industry), or want to chat about this work, please reach out.
A focus on Anna's preprint, but several other updates too, including Faith Adegoke joining us to work on AMR and several other preprints
bigdatabiology.substack.com/p/bdb-lab-se...

A focus on Anna's preprint, but several other updates too, including Faith Adegoke joining us to work on AMR and several other preprints
bigdatabiology.substack.com/p/bdb-lab-se...
@bigdatabiology.bsky.social lab in Shanghai see the light! The work includes my three favorite things research-wise: 🦠 microbiome, 🧬 long-reads, and 🐶 dogs.
See our new preprint:
www.biorxiv.org/content/10.1...

@bigdatabiology.bsky.social lab in Shanghai see the light! The work includes my three favorite things research-wise: 🦠 microbiome, 🧬 long-reads, and 🐶 dogs.
See our new preprint:
www.biorxiv.org/content/10.1...
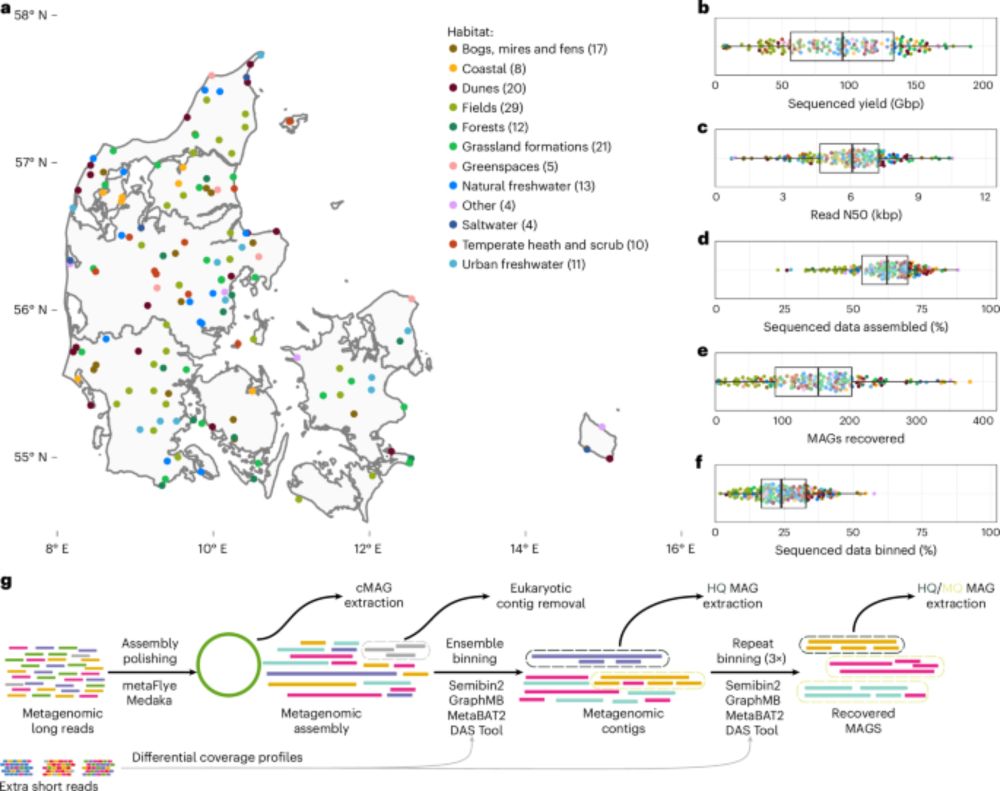
Using ONT+Illumina, we get better MAGs than to corresponding species representative in public databases
doi.org/10.1101/2025...

Using ONT+Illumina, we get better MAGs than to corresponding species representative in public databases
doi.org/10.1101/2025...

🔗 www.sciencedirect.com/science/arti...

🔗 www.sciencedirect.com/science/arti...
Let's talk about Microbiome! The speakers are will be live for Q&A.
Join at: cassyni.com/s/mvif-3
Today's backstage team:
@aroneys.bsky.social
@annacusco.bsky.social
@pamferretti.bsky.social
@azufre451.bsky.social
@kruthirao.bsky.social
@JoseCaparros
Let's talk about Microbiome! The speakers are will be live for Q&A.
Join at: cassyni.com/s/mvif-3
Today's backstage team:
@aroneys.bsky.social
@annacusco.bsky.social
@pamferretti.bsky.social
@azufre451.bsky.social
@kruthirao.bsky.social
@JoseCaparros
Fully funded studentships available to work on a range of topics, from small proteins to developing computational tools to study the global microbiome
Fully funded studentships available to work on a range of topics, from small proteins to developing computational tools to study the global microbiome
@sjmcilroy.bsky.social, @benjwoodcroft.bsky.social, @jamesvolmer.bsky.social
doi.org/10.21203/rs....
🧵1/6
@sjmcilroy.bsky.social, @benjwoodcroft.bsky.social, @jamesvolmer.bsky.social
doi.org/10.21203/rs....
🧵1/6
...and #MVIF 34 program is out! 🤩
⭐️MicroTalks:
🇹🇼 Ha T. Doan
🇦🇺 Vedanth Ramji
⭐️ Keynote:
🇺🇸 Sean Gibbons @gibbological.bsky.social
⭐️Selected talks:
🇯🇵 Yuya Kiguchi
🇦🇺 Kurtis Budden
🇨🇭 Amit Halkhoree (#Roche)
Registration: cassyni.com/s/mvif-34

...and #MVIF 34 program is out! 🤩
⭐️MicroTalks:
🇹🇼 Ha T. Doan
🇦🇺 Vedanth Ramji
⭐️ Keynote:
🇺🇸 Sean Gibbons @gibbological.bsky.social
⭐️Selected talks:
🇯🇵 Yuya Kiguchi
🇦🇺 Kurtis Budden
🇨🇭 Amit Halkhoree (#Roche)
Registration: cassyni.com/s/mvif-34
Check out these people below and please let me know if you would like to be included!
go.bsky.app/6vPBEty
Check out these people below and please let me know if you would like to be included!
go.bsky.app/6vPBEty
🦠🧫🔬
Still learning to use bluesky and might have missed you, let me know if you would like to be included.
go.bsky.app/Fq36egy
🦠🧫🔬
Still learning to use bluesky and might have missed you, let me know if you would like to be included.
go.bsky.app/Fq36egy
If you're on it and don't want to be I will happily remove you
go.bsky.app/H5k2p2g
If you're on it and don't want to be I will happily remove you
go.bsky.app/H5k2p2g
I've been thinking about this for a few years: the incentives in the field are to produce false positives
luispedro.substack.com/p/why-are-bi...
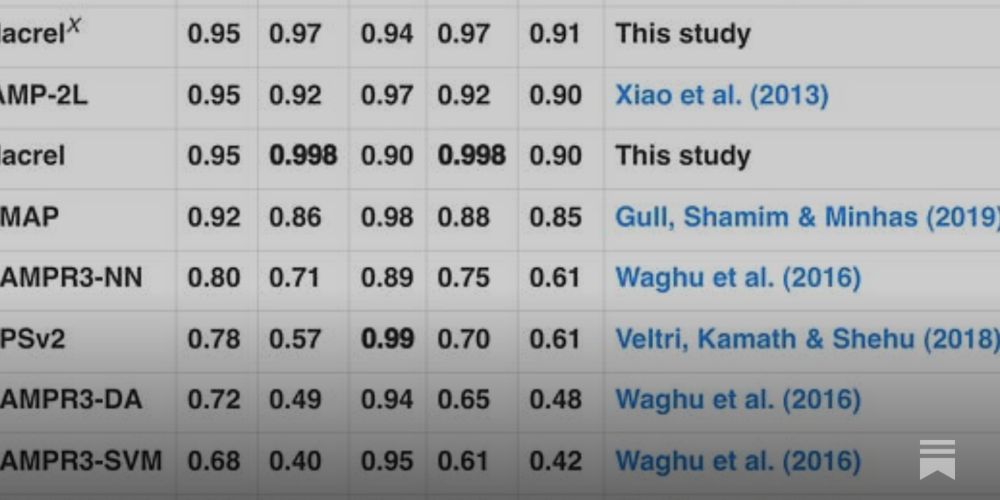
I've been thinking about this for a few years: the incentives in the field are to produce false positives
luispedro.substack.com/p/why-are-bi...
the argNorm preprint is now available on #QUT ePrints: eprints.qut.edu.au/252448
We* designed a tool for normalizing ARG annotations across currently popular tools & dbs.
*@svetlanaup.bsky.social
Vedanth Ramji
Hui Chong
Yiqian Duan
Finlay Maguire
@luispedrocoelho.bsky.social
1/6
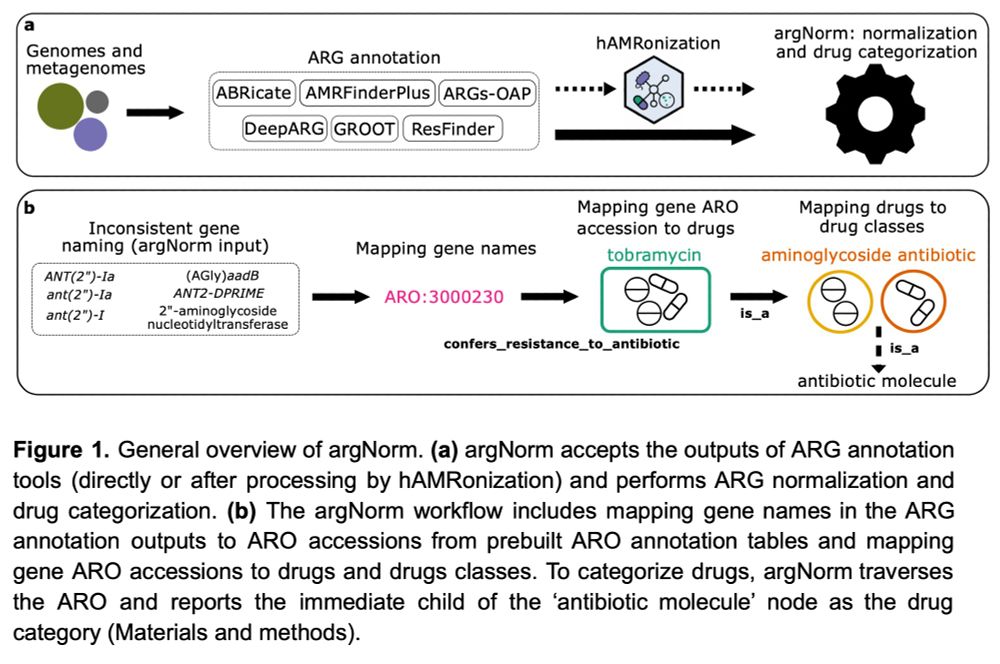
the argNorm preprint is now available on #QUT ePrints: eprints.qut.edu.au/252448
We* designed a tool for normalizing ARG annotations across currently popular tools & dbs.
*@svetlanaup.bsky.social
Vedanth Ramji
Hui Chong
Yiqian Duan
Finlay Maguire
@luispedrocoelho.bsky.social
1/6

Everyone is welcome!

Everyone is welcome!
As part of our ongoing efforts to understand small proteins in prokaryotes, we catalogued almost 1 billion sequences!
www.biorxiv.org/content/10.1...

As part of our ongoing efforts to understand small proteins in prokaryotes, we catalogued almost 1 billion sequences!
www.biorxiv.org/content/10.1...
New paper in Science shows this is achieved by nutrient blocking -- diverse communities will consume all the nutrients an incoming pathogen needs to colonize
www.science.org/doi/10.1126/...

New paper in Science shows this is achieved by nutrient blocking -- diverse communities will consume all the nutrients an incoming pathogen needs to colonize
www.science.org/doi/10.1126/...
This is a little tool to map ARG (antibiotic resistance gene) annotations from different tools to the same common ontology (ARO from CARD)

This is a little tool to map ARG (antibiotic resistance gene) annotations from different tools to the same common ontology (ARO from CARD)

