
Anna Cusco
@annacusco.bsky.social
Microbiome scientist | Metagenomics | Long-read sequencing
Postdoc at Big Data Biology Lab
Postdoc at Big Data Biology Lab
Reposted by Anna Cusco
If you made it this far, thanks for reading. I hope you enjoyed it.
I am currently looking for my next research adventure. If you have insights on the microbiome/microbial genomics job market in Europe (academia & industry), or want to chat about this work, please reach out.
I am currently looking for my next research adventure. If you have insights on the microbiome/microbial genomics job market in Europe (academia & industry), or want to chat about this work, please reach out.


September 23, 2025 at 11:24 AM
If you made it this far, thanks for reading. I hope you enjoyed it.
I am currently looking for my next research adventure. If you have insights on the microbiome/microbial genomics job market in Europe (academia & industry), or want to chat about this work, please reach out.
I am currently looking for my next research adventure. If you have insights on the microbiome/microbial genomics job market in Europe (academia & industry), or want to chat about this work, please reach out.
That sounds great, thanks for sharing!
September 25, 2025 at 10:05 AM
That sounds great, thanks for sharing!
If you made it this far, thanks for reading. I hope you enjoyed it.
I am currently looking for my next research adventure. If you have insights on the microbiome/microbial genomics job market in Europe (academia & industry), or want to chat about this work, please reach out.
I am currently looking for my next research adventure. If you have insights on the microbiome/microbial genomics job market in Europe (academia & industry), or want to chat about this work, please reach out.


September 23, 2025 at 11:24 AM
If you made it this far, thanks for reading. I hope you enjoyed it.
I am currently looking for my next research adventure. If you have insights on the microbiome/microbial genomics job market in Europe (academia & industry), or want to chat about this work, please reach out.
I am currently looking for my next research adventure. If you have insights on the microbiome/microbial genomics job market in Europe (academia & industry), or want to chat about this work, please reach out.
I would like to thank all co-authors: Yiqian Duan, Fernando Gil, Alexei Chklovski, Nithya Kruthi, Shaojun Pan, Sofia Forslund, Susanne Lau, Ulrike Löber, Xing-Ming Zhao, and especially @luispedrocoelho.bsky.social
And of course, all the dog owners and the main characters of this story🐶
And of course, all the dog owners and the main characters of this story🐶

September 23, 2025 at 11:24 AM
I would like to thank all co-authors: Yiqian Duan, Fernando Gil, Alexei Chklovski, Nithya Kruthi, Shaojun Pan, Sofia Forslund, Susanne Lau, Ulrike Löber, Xing-Ming Zhao, and especially @luispedrocoelho.bsky.social
And of course, all the dog owners and the main characters of this story🐶
And of course, all the dog owners and the main characters of this story🐶
All the generated data and resources are publicly available.
You can play around with the Shanghai dog MAG catalog here: sh-dog-mags.big-data-biology.org
where we added MAG basic information, ARGs annotation, and linked the 16S rRNA genes to MicrobeAtlas.
More data at Zenodo & ENA.
You can play around with the Shanghai dog MAG catalog here: sh-dog-mags.big-data-biology.org
where we added MAG basic information, ARGs annotation, and linked the 16S rRNA genes to MicrobeAtlas.
More data at Zenodo & ENA.

September 23, 2025 at 11:24 AM
All the generated data and resources are publicly available.
You can play around with the Shanghai dog MAG catalog here: sh-dog-mags.big-data-biology.org
where we added MAG basic information, ARGs annotation, and linked the 16S rRNA genes to MicrobeAtlas.
More data at Zenodo & ENA.
You can play around with the Shanghai dog MAG catalog here: sh-dog-mags.big-data-biology.org
where we added MAG basic information, ARGs annotation, and linked the 16S rRNA genes to MicrobeAtlas.
More data at Zenodo & ENA.
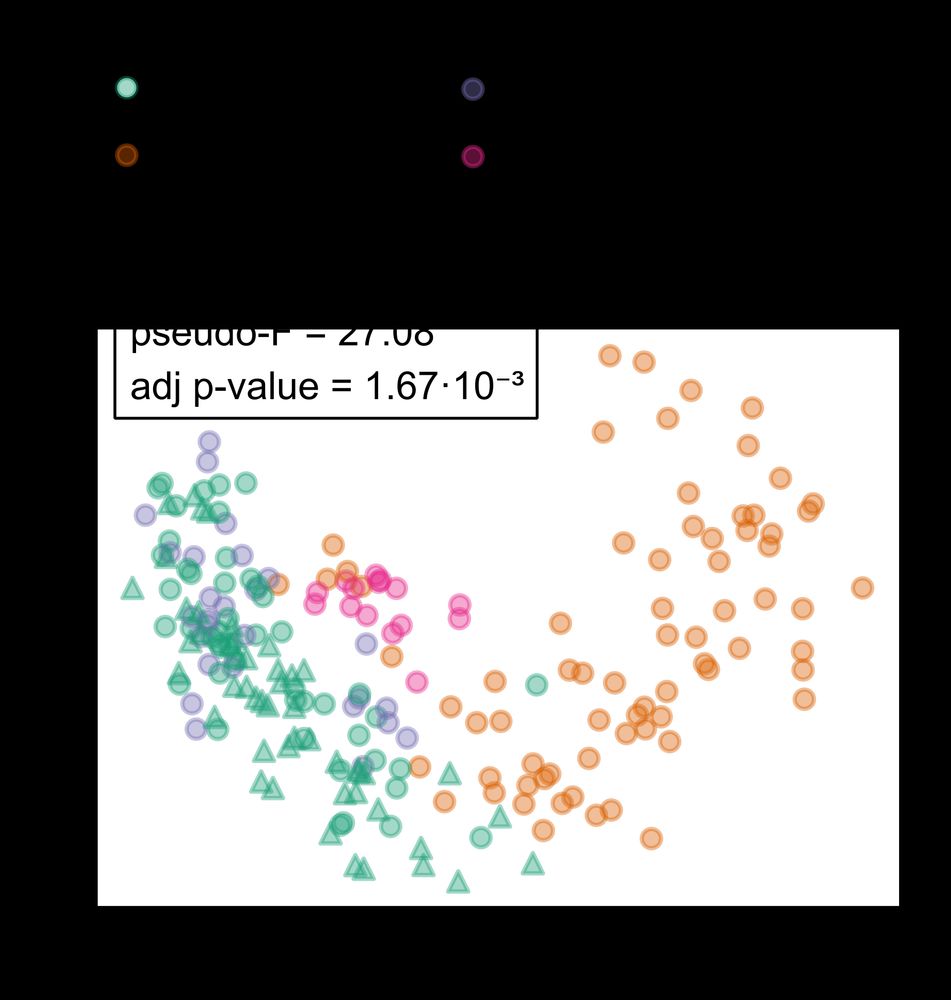
Using global dog gut microbiome data, we found that the living environment (household, colony, free-roaming) was the strongest factor shaping the gut microbiome composition.
(This is consistent with the mapping results: higher mapping rates for pet dog vs. non-pet dog cohorts)
(This is consistent with the mapping results: higher mapping rates for pet dog vs. non-pet dog cohorts)

September 23, 2025 at 11:24 AM
Using global dog gut microbiome data, we found that the living environment (household, colony, free-roaming) was the strongest factor shaping the gut microbiome composition.
(This is consistent with the mapping results: higher mapping rates for pet dog vs. non-pet dog cohorts)
(This is consistent with the mapping results: higher mapping rates for pet dog vs. non-pet dog cohorts)
In general, our species-level MAG representatives were more contiguous and had a higher quality than the reference genome assembly in public databases —especially regarding the presence of rRNA and mobile genetic elements, which are often missed in short-read assemblies.

September 23, 2025 at 11:24 AM
In general, our species-level MAG representatives were more contiguous and had a higher quality than the reference genome assembly in public databases —especially regarding the presence of rRNA and mobile genetic elements, which are often missed in short-read assemblies.
Our Shanghai dog catalogs proved to be globally representative🌍: over 90% of reads (median value) from pet dog cohorts in Germany, South Africa, and the USA mapped to them.

September 23, 2025 at 11:24 AM
Our Shanghai dog catalogs proved to be globally representative🌍: over 90% of reads (median value) from pet dog cohorts in Germany, South Africa, and the USA mapped to them.
Let's go to some of the main messages:
🧬We recovered 2,676 MAGs from Shanghai dogs, ~72% were near-finished (high-quality regarding MIMAG criteria) & highly contiguous.
⭕We recovered 185 circular extrachromosomal elements like plasmids and viruses from the same dogs.
🧬We recovered 2,676 MAGs from Shanghai dogs, ~72% were near-finished (high-quality regarding MIMAG criteria) & highly contiguous.
⭕We recovered 185 circular extrachromosomal elements like plasmids and viruses from the same dogs.


September 23, 2025 at 11:24 AM
Let's go to some of the main messages:
🧬We recovered 2,676 MAGs from Shanghai dogs, ~72% were near-finished (high-quality regarding MIMAG criteria) & highly contiguous.
⭕We recovered 185 circular extrachromosomal elements like plasmids and viruses from the same dogs.
🧬We recovered 2,676 MAGs from Shanghai dogs, ~72% were near-finished (high-quality regarding MIMAG criteria) & highly contiguous.
⭕We recovered 185 circular extrachromosomal elements like plasmids and viruses from the same dogs.
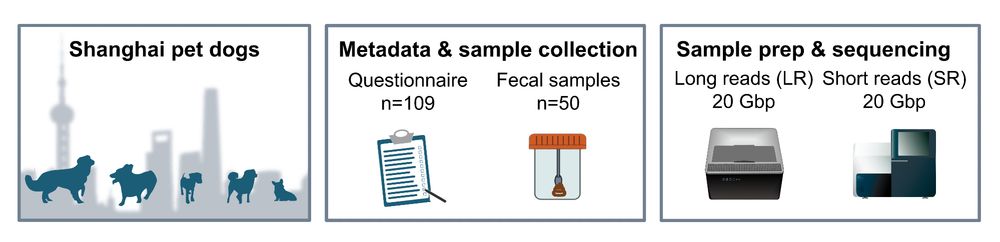
Each stool sample was deeply sequenced 🐶💩: 20 Gbp Illumina short-reads + (at least) 20 Gbp Nanopore long-reads per dog. That's a substantial throughput for assembling MAGs from complex metagenomes🧬!
September 23, 2025 at 11:24 AM
Each stool sample was deeply sequenced 🐶💩: 20 Gbp Illumina short-reads + (at least) 20 Gbp Nanopore long-reads per dog. That's a substantial throughput for assembling MAGs from complex metagenomes🧬!
We started by sharing questionnaires via WeChat 📲 and then collected 💩 samples from 51 pet dogs across Shanghai 🐶. Along the way, we met many doggies (& their humans)!

September 23, 2025 at 11:24 AM
We started by sharing questionnaires via WeChat 📲 and then collected 💩 samples from 51 pet dogs across Shanghai 🐶. Along the way, we met many doggies (& their humans)!
If you made it this far, thanks for reading. I hope you enjoyed the thread.
I am currently looking for my next research adventure. If you have insights on the microbiome/microbial genomics🧬🦠 job market in Europe (academia and industry), or want to chat about this work, please reach out!
I am currently looking for my next research adventure. If you have insights on the microbiome/microbial genomics🧬🦠 job market in Europe (academia and industry), or want to chat about this work, please reach out!


September 23, 2025 at 10:42 AM
If you made it this far, thanks for reading. I hope you enjoyed the thread.
I am currently looking for my next research adventure. If you have insights on the microbiome/microbial genomics🧬🦠 job market in Europe (academia and industry), or want to chat about this work, please reach out!
I am currently looking for my next research adventure. If you have insights on the microbiome/microbial genomics🧬🦠 job market in Europe (academia and industry), or want to chat about this work, please reach out!
I would like to thank all co-authors: Yiqian Duan, Fernando Gil, Alexei Chklovski, Nithya Kruthi, Shaojun Pan, Sofia Forslund, Susanne Lau, Ulrike Löber, Xing-Ming Zhao, and especially @luispedrocoelho.bsky.social
And of course, all the dog owners and the main characters of this story 🐶
And of course, all the dog owners and the main characters of this story 🐶

September 23, 2025 at 10:42 AM
I would like to thank all co-authors: Yiqian Duan, Fernando Gil, Alexei Chklovski, Nithya Kruthi, Shaojun Pan, Sofia Forslund, Susanne Lau, Ulrike Löber, Xing-Ming Zhao, and especially @luispedrocoelho.bsky.social
And of course, all the dog owners and the main characters of this story 🐶
And of course, all the dog owners and the main characters of this story 🐶
All the generated data and resources are publicly available.
You can play around with the Shanghai dog MAG catalog here: sh-dog-mags.big-data-biology.org
where we have added MAG basic information, ARGs annotation, and linked 16S rRNA genes to MicrobeAtlas.
More data at Zenodo and ENA.
You can play around with the Shanghai dog MAG catalog here: sh-dog-mags.big-data-biology.org
where we have added MAG basic information, ARGs annotation, and linked 16S rRNA genes to MicrobeAtlas.
More data at Zenodo and ENA.

September 23, 2025 at 10:42 AM
All the generated data and resources are publicly available.
You can play around with the Shanghai dog MAG catalog here: sh-dog-mags.big-data-biology.org
where we have added MAG basic information, ARGs annotation, and linked 16S rRNA genes to MicrobeAtlas.
More data at Zenodo and ENA.
You can play around with the Shanghai dog MAG catalog here: sh-dog-mags.big-data-biology.org
where we have added MAG basic information, ARGs annotation, and linked 16S rRNA genes to MicrobeAtlas.
More data at Zenodo and ENA.
Using global dog gut microbiome data, we found that the living environment (household, colony, free-roaming) was the strongest factor shaping the gut microbiome composition.
(This is consistent with previous mapping results to our catalog: higher mapping rates for pet dog vs. non-pet dog cohorts)
(This is consistent with previous mapping results to our catalog: higher mapping rates for pet dog vs. non-pet dog cohorts)
September 23, 2025 at 10:42 AM
Using global dog gut microbiome data, we found that the living environment (household, colony, free-roaming) was the strongest factor shaping the gut microbiome composition.
(This is consistent with previous mapping results to our catalog: higher mapping rates for pet dog vs. non-pet dog cohorts)
(This is consistent with previous mapping results to our catalog: higher mapping rates for pet dog vs. non-pet dog cohorts)
In general, our species-level MAG representatives were more contiguous and had a higher quality than the reference genome assembly in public databases —especially regarding the presence of rRNA and mobile genetic elements, which are often missed in short-read assemblies.

September 23, 2025 at 10:42 AM
In general, our species-level MAG representatives were more contiguous and had a higher quality than the reference genome assembly in public databases —especially regarding the presence of rRNA and mobile genetic elements, which are often missed in short-read assemblies.
Our Shanghai dog catalogs proved to be globally representative🌍: over 90% of reads (median value) from pet dog cohorts in Germany, South Africa, and the USA mapped to them.

September 23, 2025 at 10:42 AM
Our Shanghai dog catalogs proved to be globally representative🌍: over 90% of reads (median value) from pet dog cohorts in Germany, South Africa, and the USA mapped to them.
Let's go to some of the main messages:
🧬We recovered 2,676 MAGs from Shanghai dogs, ~72% were near-finished (high-quality regarding MIMAG criteria) and highly contiguous.
⭕Additionally, we retrieved 185 circular extrachromosomal elements like plasmids and viruses from the same dogs.
🧬We recovered 2,676 MAGs from Shanghai dogs, ~72% were near-finished (high-quality regarding MIMAG criteria) and highly contiguous.
⭕Additionally, we retrieved 185 circular extrachromosomal elements like plasmids and viruses from the same dogs.


September 23, 2025 at 10:42 AM
Let's go to some of the main messages:
🧬We recovered 2,676 MAGs from Shanghai dogs, ~72% were near-finished (high-quality regarding MIMAG criteria) and highly contiguous.
⭕Additionally, we retrieved 185 circular extrachromosomal elements like plasmids and viruses from the same dogs.
🧬We recovered 2,676 MAGs from Shanghai dogs, ~72% were near-finished (high-quality regarding MIMAG criteria) and highly contiguous.
⭕Additionally, we retrieved 185 circular extrachromosomal elements like plasmids and viruses from the same dogs.
Each stool sample was deeply sequenced 🐶💩: 20 Gbp Illumina short-reads + (at least) 20 Gbp Nanopore long-reads per dog. That's a substantial throughput for assembling MAGs from complex metagenomes🧬!

September 23, 2025 at 10:42 AM
Each stool sample was deeply sequenced 🐶💩: 20 Gbp Illumina short-reads + (at least) 20 Gbp Nanopore long-reads per dog. That's a substantial throughput for assembling MAGs from complex metagenomes🧬!
We started by sharing questionnaires via WeChat 📲 and then collected 💩 samples from 51 pet dogs across Shanghai 🐶. Along the way, we met many doggies (& their humans)!

September 23, 2025 at 10:42 AM
We started by sharing questionnaires via WeChat 📲 and then collected 💩 samples from 51 pet dogs across Shanghai 🐶. Along the way, we met many doggies (& their humans)!

