



💻🧬El BSC intenta resolver algunos de los grandes desafíos médicos. Su departamento de Ciencias de la Vida trabaja en #cáncer, patologías raras, nuevos fármacos o análisis genómico.

💻🧬El BSC intenta resolver algunos de los grandes desafíos médicos. Su departamento de Ciencias de la Vida trabaja en #cáncer, patologías raras, nuevos fármacos o análisis genómico.
It turned into multi-dimensional puzzles and alignement problems that we really enjoyed solving! Congrats to Daniel Lepe-Soltero, the postdoc who led this work.
Link -> arxiv.org/abs/2503.18856

It turned into multi-dimensional puzzles and alignement problems that we really enjoyed solving! Congrats to Daniel Lepe-Soltero, the postdoc who led this work.
Link -> arxiv.org/abs/2503.18856
We solve a long-standing problem in the field: how to perform data integration for small unpaired datasets
Our solution: learning with class imbalance on both reference and target datasets!

We solve a long-standing problem in the field: how to perform data integration for small unpaired datasets
Our solution: learning with class imbalance on both reference and target datasets!
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...

🧬 When you only have a few datasets available for multi-omics integration, for instance because you work on rare diseases, one solution is Transfer Learning. Introducing #MOTL - now available in Genome Biology!
doi.org/10.1186/s130...
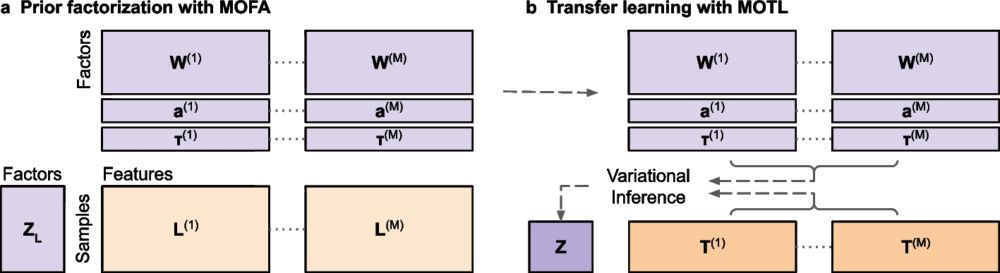
🧬 When you only have a few datasets available for multi-omics integration, for instance because you work on rare diseases, one solution is Transfer Learning. Introducing #MOTL - now available in Genome Biology!
doi.org/10.1186/s130...
Interested and attending #ISMBECCB2025? Let’s connect!
🔗 www.jobbnorge.no/en/available...

Interested and attending #ISMBECCB2025? Let’s connect!
🔗 www.jobbnorge.no/en/available...
Congrats:
1: Alejandra Paulina Perez Gonzalez
2. Michael Costanzo
3. Jong Chan Lim
👏👏👏 Exciting to have Alejandra and Jong in the audience and congrats to Michael!
Congrats:
1: Alejandra Paulina Perez Gonzalez
2. Michael Costanzo
3. Jong Chan Lim
👏👏👏 Exciting to have Alejandra and Jong in the audience and congrats to Michael!
@fionabrinkman.bsky.social is talking about "Microbiome multitudes and metadata madness" connecting the #Microbiome and #NetworkBiology communities!
#ISMBECCB2025

@fionabrinkman.bsky.social is talking about "Microbiome multitudes and metadata madness" connecting the #Microbiome and #NetworkBiology communities!
#ISMBECCB2025
#ISMBECCB2025 #NetworkBiology

#ISMBECCB2025 #NetworkBiology
Dongmin Bang is presenting his proceeding paper on "MixingDTA: Improved Drug-Target Affinity Prediction by Extending Mixup with Guilt-By-Association"
doi.org/10.1093/bioi...
#ISMBECCB2025 #NetworkBiology

Dongmin Bang is presenting his proceeding paper on "MixingDTA: Improved Drug-Target Affinity Prediction by Extending Mixup with Guilt-By-Association"
doi.org/10.1093/bioi...
#ISMBECCB2025 #NetworkBiology

He is sharing his research on "Improving Target-Adverse Event Association Prediction by Mitigating Topological Imbalance in Knowledge Graphs" with the #NetBio audience! ✨
#ISMBECCB2025 #NetworkBiology #KnowledgeGraphs

He is sharing his research on "Improving Target-Adverse Event Association Prediction by Mitigating Topological Imbalance in Knowledge Graphs" with the #NetBio audience! ✨
#ISMBECCB2025 #NetworkBiology #KnowledgeGraphs
nf-co.re/diseasemodul...
github.com/nf-core/dise...
#ISMBECCB2025 #NetworkBiology #NetBio

nf-co.re/diseasemodul...
github.com/nf-core/dise...
#ISMBECCB2025 #NetworkBiology #NetBio
Samuele Firmani is presenting GATE (Graph Antiviral Target Explorer) to predict disease genes in viral infections with Message Passing Neural Networks"
#ISMBECCB2025 #NetworkBiology
Samuele Firmani is presenting GATE (Graph Antiviral Target Explorer) to predict disease genes in viral infections with Message Passing Neural Networks"
#ISMBECCB2025 #NetworkBiology
Erik Sonnhammer presents #FunCoup6, an upgrade to the functional association network database, now with enhanced regulatory links, a redesigned interface, and powerful new tools for comparative interactomics and disease module analysis.
funcoup.org
#ISMBECCB2025 #NetworkBiology

Erik Sonnhammer presents #FunCoup6, an upgrade to the functional association network database, now with enhanced regulatory links, a redesigned interface, and powerful new tools for comparative interactomics and disease module analysis.
funcoup.org
#ISMBECCB2025 #NetworkBiology
Dewei Hu presents:
SPACE: STRING proteins as complementary embeddings
Cross-species network embeddings meet protein function prediction, leveraging STRING across 1,300+ eukaryotes. A powerful complement to sequence embeddings!
#NetworkBiology
Dewei Hu presents:
SPACE: STRING proteins as complementary embeddings
Cross-species network embeddings meet protein function prediction, leveraging STRING across 1,300+ eukaryotes. A powerful complement to sequence embeddings!
#NetworkBiology
✨🔊We are excited to welcome our 2nd keynote speaker Jan Baumbach talking about "Quantum computing for network medicine-based epistatic disease mechanism mining - Fake it till you make it?" ✨
#ISMBECCB2025 #NetworkBiology

✨🔊We are excited to welcome our 2nd keynote speaker Jan Baumbach talking about "Quantum computing for network medicine-based epistatic disease mechanism mining - Fake it till you make it?" ✨
#ISMBECCB2025 #NetworkBiology
#ISMBECCB2025 #NetworkBiology

#ISMBECCB2025 #NetworkBiology
Lucas Gillenwater is presenting his #ISMBECCB2025 proceeding paper "GRACKLE: An interpretable matrix factorization approach for biomedical representation learning".
doi.org/10.1093/bioi...
#NetworkBiology

Lucas Gillenwater is presenting his #ISMBECCB2025 proceeding paper "GRACKLE: An interpretable matrix factorization approach for biomedical representation learning".
doi.org/10.1093/bioi...
#NetworkBiology
@beatrizurda.bsky.social: Disentangling the genetic and non-genetic origin of disease co-occurrences
#ISMBECCB2025 #NetworkBiology

@beatrizurda.bsky.social: Disentangling the genetic and non-genetic origin of disease co-occurrences
#ISMBECCB2025 #NetworkBiology
#ISMBECCB2025 #NetworkBiology

#ISMBECCB2025 #NetworkBiology
github.com/ibrahimsagga...
#ISMBECCB2025 #NetworkBiology

github.com/ibrahimsagga...
#ISMBECCB2025 #NetworkBiology

