
Albert Dominguez Mantes
@albertdm.bsky.social
PhD student @ EPFL in computer vision for microscopy | Prev Data Science @ UPC
Pinned
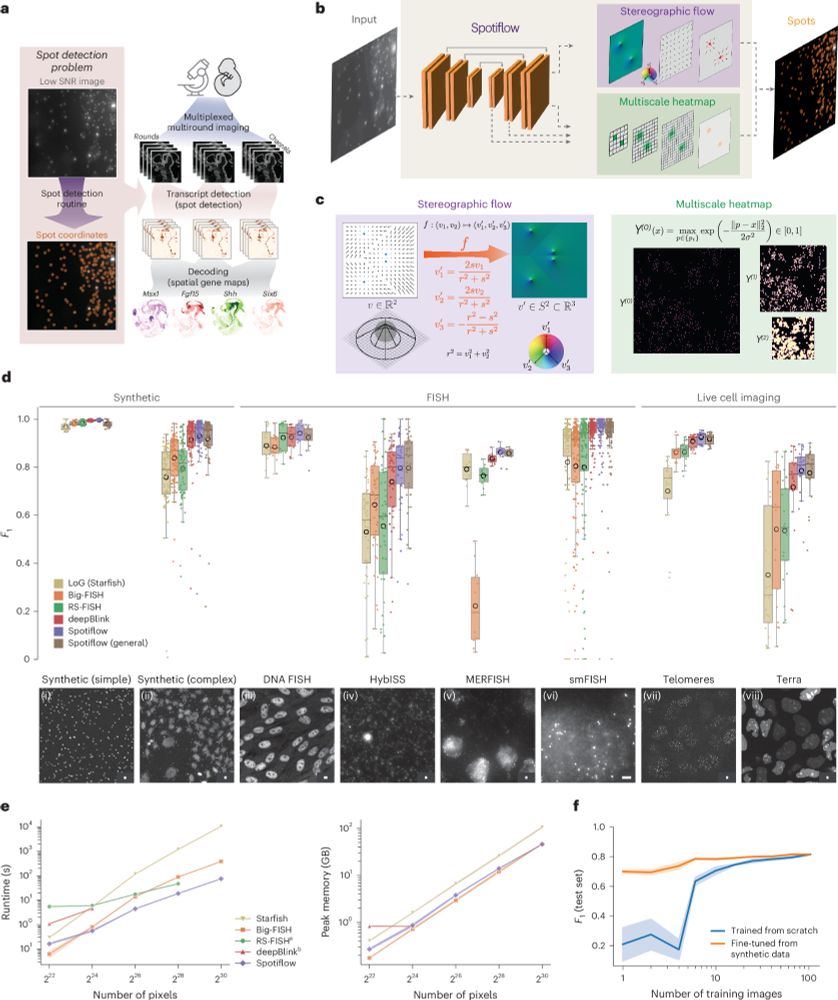
Spotiflow: accurate and efficient spot detection for fluorescence microscopy with deep stereographic flow regression
Nature Methods - Spotiflow uses deep learning for subpixel-accurate spot detection in diverse 2D and 3D images. The improved accuracy offered by Spotiflow enables improved biological insights in...
rdcu.be
Spotiflow, our deep learning based spot detection method for microscopy, is now published in @natmethods.nature.com!
Since the pre-print, we have added many features, notably native 3D detection!
@maweigert.bsky.social @gioelelamanno.bsky.social @epfl-brainmind.bsky.social
Paper: rdcu.be/epIB7
(1/N)
Since the pre-print, we have added many features, notably native 3D detection!
@maweigert.bsky.social @gioelelamanno.bsky.social @epfl-brainmind.bsky.social
Paper: rdcu.be/epIB7
(1/N)
Reposted by Albert Dominguez Mantes
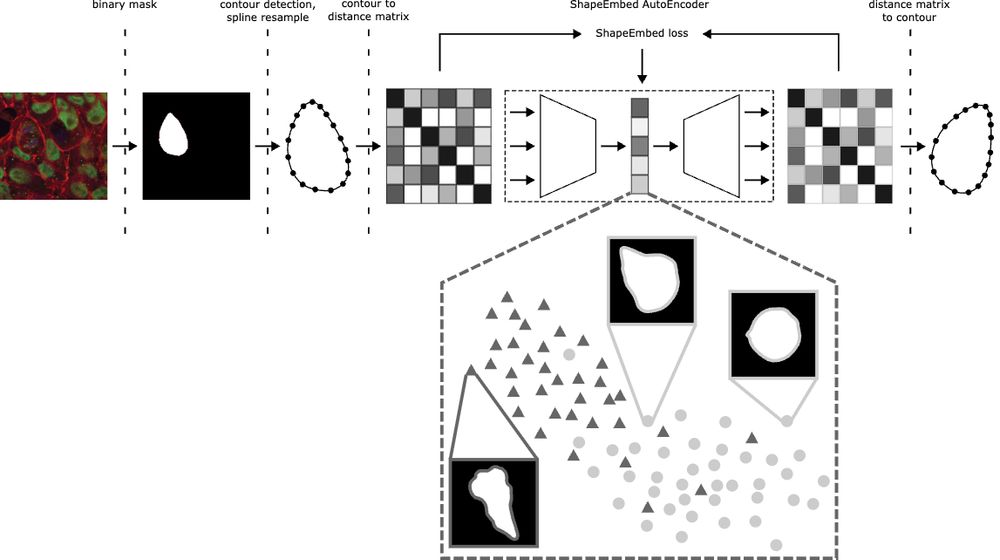
We’ve upgraded ShapeEmbed 🎉 ShapeEmbedLite decodes latent codes via an MLP to guarantee valid EDMs, making it lighter and ideal for small microscopy datasets or limited compute. Hear more at my BIC workshop talk or poster at #ICCV2025! Try it out at github.com/uhlmanngroup...

October 17, 2025 at 7:35 AM
We’ve upgraded ShapeEmbed 🎉 ShapeEmbedLite decodes latent codes via an MLP to guarantee valid EDMs, making it lighter and ideal for small microscopy datasets or limited compute. Hear more at my BIC workshop talk or poster at #ICCV2025! Try it out at github.com/uhlmanngroup...
Reposted by Albert Dominguez Mantes
🧠 The Lipid #Brain Atlas is out now! If you think #lipids are boring and membranes are all the same, prepare to be surprised. Led by @lucafusarbassini.bsky.social with Giovanni D'Angelo's lab, we mapped membrane lipids in the mouse brain at high resolution.
www.biorxiv.org/cgi/content/...
www.biorxiv.org/cgi/content/...

October 16, 2025 at 6:23 AM
🧠 The Lipid #Brain Atlas is out now! If you think #lipids are boring and membranes are all the same, prepare to be surprised. Led by @lucafusarbassini.bsky.social with Giovanni D'Angelo's lab, we mapped membrane lipids in the mouse brain at high resolution.
www.biorxiv.org/cgi/content/...
www.biorxiv.org/cgi/content/...
Reposted by Albert Dominguez Mantes
You may have heard me talk about learned shape representations for a long time, but it took @afoix.bsky.social's hard work to bring it to life 🍾 Catch her at @neuripsconf.bsky.social to find out more!!
Happy to share that ShapeEmbed has been accepted at @neuripsconf.bsky.social 🎉 SE is self-supervised framework to encode 2D contours from microscopy & natural images into a latent representation invariant to translation, scaling, rotation, reflection & point indexing
📄 arxiv.org/pdf/2507.01009 (1/N)
📄 arxiv.org/pdf/2507.01009 (1/N)
September 23, 2025 at 8:57 AM
You may have heard me talk about learned shape representations for a long time, but it took @afoix.bsky.social's hard work to bring it to life 🍾 Catch her at @neuripsconf.bsky.social to find out more!!
Reposted by Albert Dominguez Mantes
Happy to share that ShapeEmbed has been accepted at @neuripsconf.bsky.social 🎉 SE is self-supervised framework to encode 2D contours from microscopy & natural images into a latent representation invariant to translation, scaling, rotation, reflection & point indexing
📄 arxiv.org/pdf/2507.01009 (1/N)
📄 arxiv.org/pdf/2507.01009 (1/N)

September 23, 2025 at 8:32 AM
Happy to share that ShapeEmbed has been accepted at @neuripsconf.bsky.social 🎉 SE is self-supervised framework to encode 2D contours from microscopy & natural images into a latent representation invariant to translation, scaling, rotation, reflection & point indexing
📄 arxiv.org/pdf/2507.01009 (1/N)
📄 arxiv.org/pdf/2507.01009 (1/N)
Reposted by Albert Dominguez Mantes
(1/14) I’m happy and proud to introduce: SpinePy – a framework to detect the "spine" of gastruloids and measure biological and physical signals in a local dynamic 3D coordinate system. www.biorxiv.org/content/10.1...
September 12, 2025 at 12:44 PM
(1/14) I’m happy and proud to introduce: SpinePy – a framework to detect the "spine" of gastruloids and measure biological and physical signals in a local dynamic 3D coordinate system. www.biorxiv.org/content/10.1...
Reposted by Albert Dominguez Mantes
Happy to share our work on uMAIA, a framework for building metabolomic atlases from mass spectrometry imaging.
With uMAIA, we mapped the lipidome of zebrafish development, uncovering spatially organized metabolic programs .
www.nature.com/articles/s41...
#developmentalbiology #MSI #lipidtime
With uMAIA, we mapped the lipidome of zebrafish development, uncovering spatially organized metabolic programs .
www.nature.com/articles/s41...
#developmentalbiology #MSI #lipidtime

Unified mass imaging maps the lipidome of vertebrate development
Nature Methods - uMAIA is an analytical framework designed to enable the construction of metabolic atlases at high resolution using mass spectrometry imaging data.
www.nature.com
September 3, 2025 at 10:40 AM
Happy to share our work on uMAIA, a framework for building metabolomic atlases from mass spectrometry imaging.
With uMAIA, we mapped the lipidome of zebrafish development, uncovering spatially organized metabolic programs .
www.nature.com/articles/s41...
#developmentalbiology #MSI #lipidtime
With uMAIA, we mapped the lipidome of zebrafish development, uncovering spatially organized metabolic programs .
www.nature.com/articles/s41...
#developmentalbiology #MSI #lipidtime
Reposted by Albert Dominguez Mantes
A new extension! Rémy made a wrapper of @albertdm.bsky.social + @maweigert.bsky.social 's Spotiflow (2/3)
forum.image.sc/t/qupath-ext...
forum.image.sc/t/qupath-ext...

QuPath extension Spotiflow is now available
Hello @qupath team & users, We are pleased to announce a new qupath-spotiflow-extension to handle spotiflow in QuPath 🎉 ! This extension reproduces the logic behind the qupath-extension-cellpose to ...
forum.image.sc
August 12, 2025 at 3:10 PM
A new extension! Rémy made a wrapper of @albertdm.bsky.social + @maweigert.bsky.social 's Spotiflow (2/3)
forum.image.sc/t/qupath-ext...
forum.image.sc/t/qupath-ext...
Reposted by Albert Dominguez Mantes
Cell tracking is never perfect, and it's important to understand the types of errors your solution contains.
Here is my stab at this: Divisualisations in @napari.org. github.com/bentaculum/d...
You spin tracks out upwards from the playing video. Green edges are correct, FP in magenta, FN in cyan.
Here is my stab at this: Divisualisations in @napari.org. github.com/bentaculum/d...
You spin tracks out upwards from the playing video. Green edges are correct, FP in magenta, FN in cyan.
July 30, 2025 at 8:15 AM
Cell tracking is never perfect, and it's important to understand the types of errors your solution contains.
Here is my stab at this: Divisualisations in @napari.org. github.com/bentaculum/d...
You spin tracks out upwards from the playing video. Green edges are correct, FP in magenta, FN in cyan.
Here is my stab at this: Divisualisations in @napari.org. github.com/bentaculum/d...
You spin tracks out upwards from the playing video. Green edges are correct, FP in magenta, FN in cyan.
Working in Martin’s group is an amazing experience, don’t hesitate to apply and reach out if you have any questions!
Do you like developing new AI vision methods for microscopy image analysis? You love theory & implementation? 1 week left to apply for a fully funded PhD position in our lab in Dresden 🇩🇪! Topics: object detection/tracking, multimodal models & more. DM/email for details! #PhD #AcademicJobs #GPUsgoBrr
Vacancy ID 12244
www.verw.tu-dresden.de
July 10, 2025 at 8:39 PM
Working in Martin’s group is an amazing experience, don’t hesitate to apply and reach out if you have any questions!
Reposted by Albert Dominguez Mantes
Out today in @natmethods.nature.com : Spotiflow, our transcript localization method for imaging-based spatial transcriptomics. Led by amazing PhD student @albertdm.bsky.social, joint work w @gioelelamanno.bsky.social at EPFL / @scadsai.bsky.social
www.nature.com/articles/s41...
rdcu.be/epIB7
www.nature.com/articles/s41...
rdcu.be/epIB7
June 6, 2025 at 7:06 PM
Out today in @natmethods.nature.com : Spotiflow, our transcript localization method for imaging-based spatial transcriptomics. Led by amazing PhD student @albertdm.bsky.social, joint work w @gioelelamanno.bsky.social at EPFL / @scadsai.bsky.social
www.nature.com/articles/s41...
rdcu.be/epIB7
www.nature.com/articles/s41...
rdcu.be/epIB7
Reposted by Albert Dominguez Mantes
A nice advance for imaging-based spatially resolved transcriptomics from the Weigert and La Manno labs. Spotiflow uses deep learning for subpixel-accurate spot detection in diverse 2D and 3D images. www.nature.com/articles/s41...
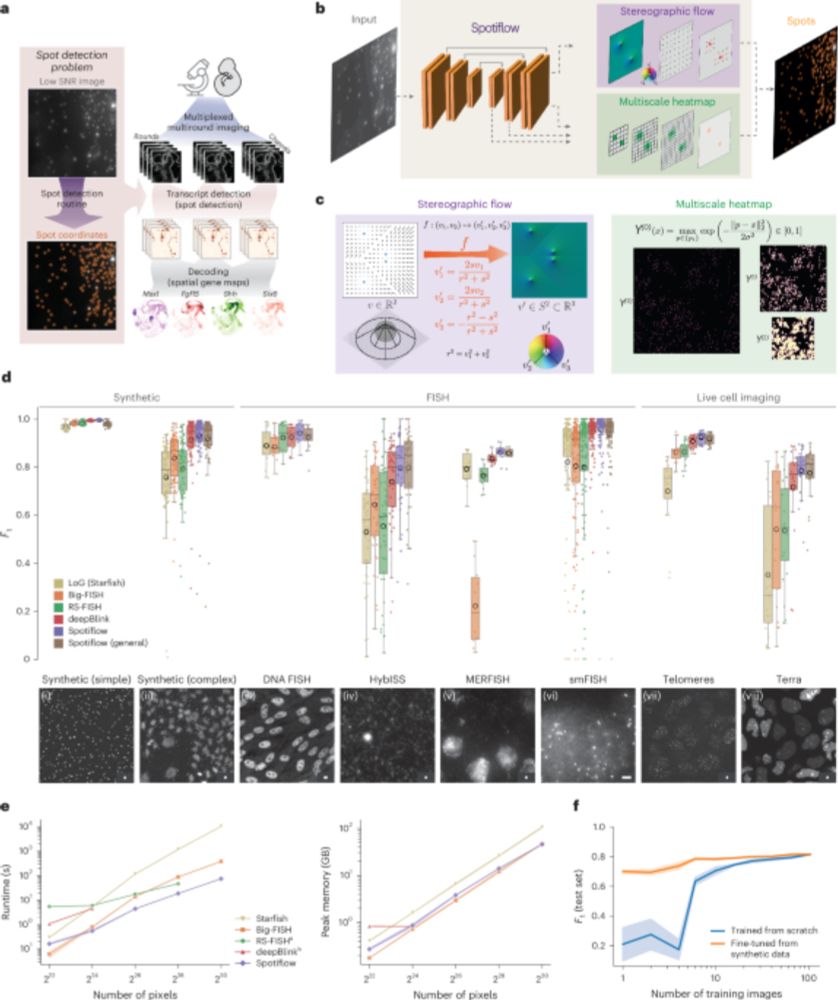
Spotiflow: accurate and efficient spot detection for fluorescence microscopy with deep stereographic flow regression - Nature Methods
Spotiflow uses deep learning for subpixel-accurate spot detection in diverse 2D and 3D images. The improved accuracy offered by Spotiflow enables improved biological insights in both iST and live imag...
www.nature.com
June 6, 2025 at 5:29 PM
A nice advance for imaging-based spatially resolved transcriptomics from the Weigert and La Manno labs. Spotiflow uses deep learning for subpixel-accurate spot detection in diverse 2D and 3D images. www.nature.com/articles/s41...
Spotiflow, our deep learning based spot detection method for microscopy, is now published in @natmethods.nature.com!
Since the pre-print, we have added many features, notably native 3D detection!
@maweigert.bsky.social @gioelelamanno.bsky.social @epfl-brainmind.bsky.social
Paper: rdcu.be/epIB7
(1/N)
Since the pre-print, we have added many features, notably native 3D detection!
@maweigert.bsky.social @gioelelamanno.bsky.social @epfl-brainmind.bsky.social
Paper: rdcu.be/epIB7
(1/N)

Spotiflow: accurate and efficient spot detection for fluorescence microscopy with deep stereographic flow regression
Nature Methods - Spotiflow uses deep learning for subpixel-accurate spot detection in diverse 2D and 3D images. The improved accuracy offered by Spotiflow enables improved biological insights in...
rdcu.be
June 6, 2025 at 6:58 PM
Spotiflow, our deep learning based spot detection method for microscopy, is now published in @natmethods.nature.com!
Since the pre-print, we have added many features, notably native 3D detection!
@maweigert.bsky.social @gioelelamanno.bsky.social @epfl-brainmind.bsky.social
Paper: rdcu.be/epIB7
(1/N)
Since the pre-print, we have added many features, notably native 3D detection!
@maweigert.bsky.social @gioelelamanno.bsky.social @epfl-brainmind.bsky.social
Paper: rdcu.be/epIB7
(1/N)
Reposted by Albert Dominguez Mantes
Get to know Spotiflow, an AI method that improves large-scale transcript localization by combining deep learning with a novel geometric detection approach.
Learn more:
👉 scads.ai/transcript-d...
Find the publication in Nature Methods:
👉 www.nature.com/articles/s41...
Learn more:
👉 scads.ai/transcript-d...
Find the publication in Nature Methods:
👉 www.nature.com/articles/s41...

AI Method Improves Transcript Detection in Microscopy Images
Spotiflow improves large-scale transcript localization by combining deep learning with a novel geometric detection approach.
scads.ai
June 6, 2025 at 10:14 AM
Get to know Spotiflow, an AI method that improves large-scale transcript localization by combining deep learning with a novel geometric detection approach.
Learn more:
👉 scads.ai/transcript-d...
Find the publication in Nature Methods:
👉 www.nature.com/articles/s41...
Learn more:
👉 scads.ai/transcript-d...
Find the publication in Nature Methods:
👉 www.nature.com/articles/s41...
Reposted by Albert Dominguez Mantes
How can AI help you to analyze your microscopy images?
Find out at the "Machine Learning for Microscopy Image Analysis" course at the @mblscience.bsky.social in Woods Hole from Aug 22-Sep 6!
Applications are due 🚨 April 30 🚨
Read on for more! 👇
www.mbl.edu/education/ad...
Find out at the "Machine Learning for Microscopy Image Analysis" course at the @mblscience.bsky.social in Woods Hole from Aug 22-Sep 6!
Applications are due 🚨 April 30 🚨
Read on for more! 👇
www.mbl.edu/education/ad...

AI@MBL: Machine Learning for Microscopy Image Analysis | Marine Biological Laboratory
The goal of this course is to familiarize researchers in the life sciences with state-of-the-art deep learning techniques for microscopy image analysis and to introduce them to tools and frameworks th...
www.mbl.edu
April 18, 2025 at 6:48 PM
How can AI help you to analyze your microscopy images?
Find out at the "Machine Learning for Microscopy Image Analysis" course at the @mblscience.bsky.social in Woods Hole from Aug 22-Sep 6!
Applications are due 🚨 April 30 🚨
Read on for more! 👇
www.mbl.edu/education/ad...
Find out at the "Machine Learning for Microscopy Image Analysis" course at the @mblscience.bsky.social in Woods Hole from Aug 22-Sep 6!
Applications are due 🚨 April 30 🚨
Read on for more! 👇
www.mbl.edu/education/ad...
Reposted by Albert Dominguez Mantes
🔬🎤 As pre-announced a few days ago... please let us proudly present to you: 𝑴𝒊𝒄𝒓𝒐𝕊𝒑𝒍𝒊𝒕 - your ticket to imaging more, imaging more gentle, and/or imaging more efficient. 🔬
doi.org/10.1101/2025...
Like ❤️, repost 🔂, and most importantly... please send feedback ✉️ our way! 🙏
doi.org/10.1101/2025...
Like ❤️, repost 🔂, and most importantly... please send feedback ✉️ our way! 🙏

February 11, 2025 at 6:02 PM
🔬🎤 As pre-announced a few days ago... please let us proudly present to you: 𝑴𝒊𝒄𝒓𝒐𝕊𝒑𝒍𝒊𝒕 - your ticket to imaging more, imaging more gentle, and/or imaging more efficient. 🔬
doi.org/10.1101/2025...
Like ❤️, repost 🔂, and most importantly... please send feedback ✉️ our way! 🙏
doi.org/10.1101/2025...
Like ❤️, repost 🔂, and most importantly... please send feedback ✉️ our way! 🙏

