

Quite an indictment of some of the current single cell "virtual cell" foundation models. Even for the relatively mundane applications, cell labeling, batch correction etc, they are poor compared to much simpler & cheaper methods.
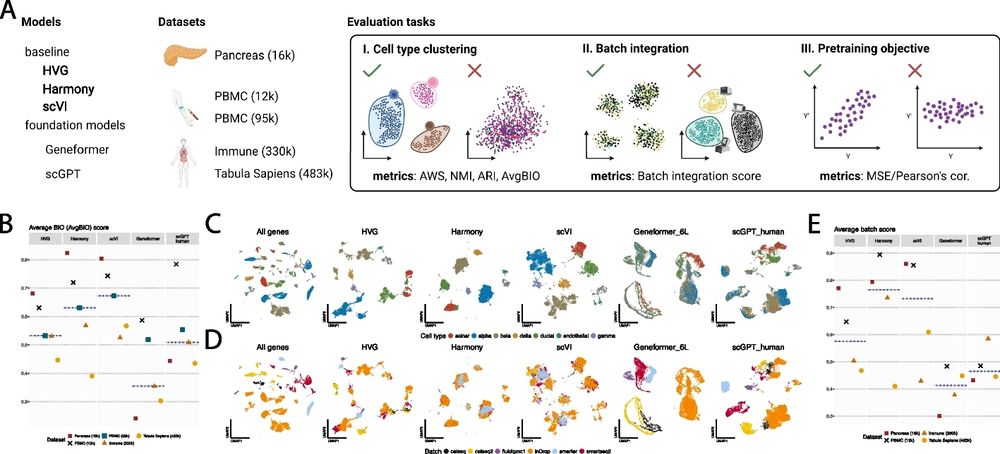
Quite an indictment of some of the current single cell "virtual cell" foundation models. Even for the relatively mundane applications, cell labeling, batch correction etc, they are poor compared to much simpler & cheaper methods.

Project led by
@nazbukina.bsky.social @zhisonghe.bsky.social @hsiuchuanlin.bsky.social

Project led by
@nazbukina.bsky.social @zhisonghe.bsky.social @hsiuchuanlin.bsky.social
doi.org/10.1101/2025...
Great work with @hsiuchuanlin.bsky.social @zhisonghe.bsky.social @graycamplab.bsky.social and Barbara Treutlein!

www.nature.com/articles/s41...

www.nature.com/articles/s41...



Find out how it can serve you ⏬
🧵1/8
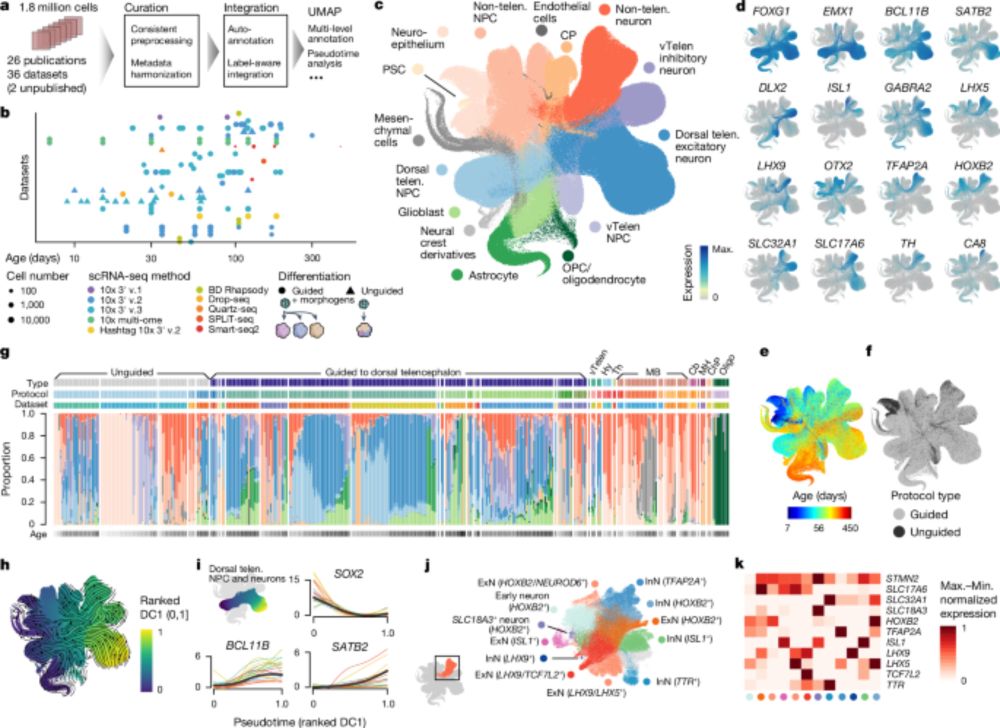
Find out how it can serve you ⏬
🧵1/8

