And a huge shoutout to the entire solab! I'm so grateful to work with these brilliant and supportive scientists every day. Keep an eye out for exciting work coming out from the team!

And a huge shoutout to the entire solab! I'm so grateful to work with these brilliant and supportive scientists every day. Keep an eye out for exciting work coming out from the team!
We're excited for all of MSA Pairformer's potential applications for biological discovery and for the future of memory and parameter efficient pLMs

We're excited for all of MSA Pairformer's potential applications for biological discovery and for the future of memory and parameter efficient pLMs



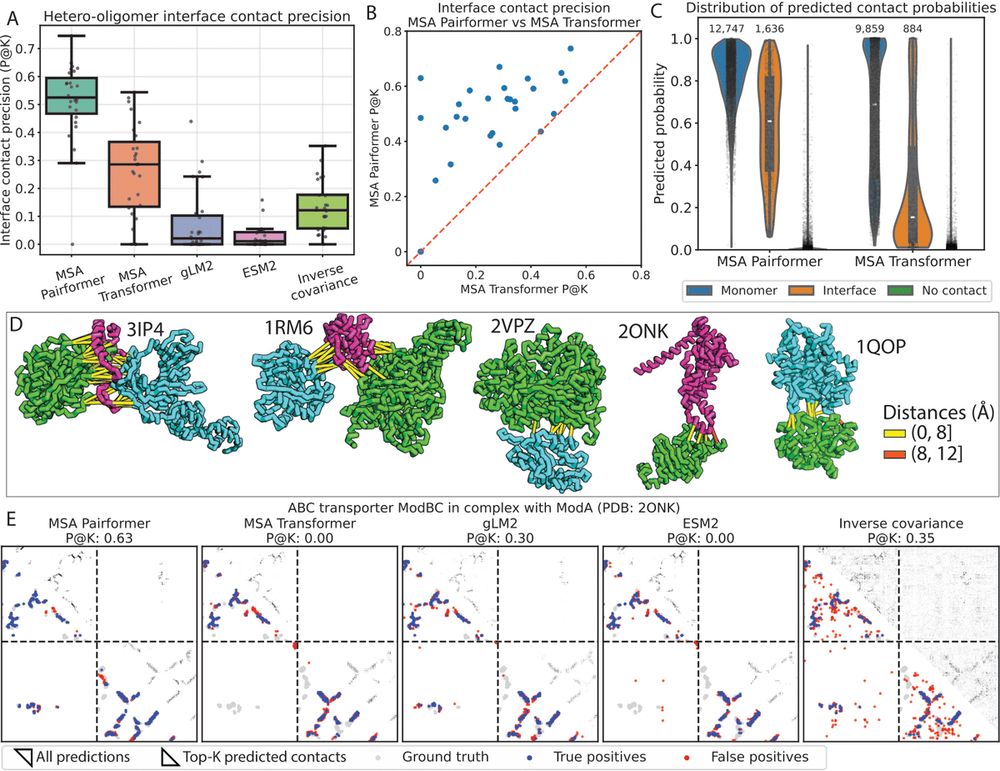
P.S. this figure slightly differs from what's in the preprint and will be updated in v2 of the paper!

P.S. this figure slightly differs from what's in the preprint and will be updated in v2 of the paper!