🧬 By charting a protein’s sequence universe, we could rationalize which versions were kept through evolution – and why many stable ones were not.
🧬 By charting a protein’s sequence universe, we could rationalize which versions were kept through evolution – and why many stable ones were not.
Thank you also to @arunsethuraman.bsky.social for handling the manuscript and @joannamasel.bsky.social for helpful comments.
academic.oup.com/g3journal/ad...
Thank you also to @arunsethuraman.bsky.social for handling the manuscript and @joannamasel.bsky.social for helpful comments.
academic.oup.com/g3journal/ad...
authors.elsevier.com/sd/article/S...
authors.elsevier.com/sd/article/S...
Aina's new preprint containing seven complete comparative allosteric maps biorxiv.org/content/10.1...
Aina's new preprint containing seven complete comparative allosteric maps biorxiv.org/content/10.1...
@xt117.bsky.social
biorxiv.org/content/10.1...

@xt117.bsky.social
biorxiv.org/content/10.1...
In our new study we cracked open the black box of early protein aggregation, and the findings could reshape how we fight neurodegeneration. 🧵👇
www.science.org/doi/10.1126/...
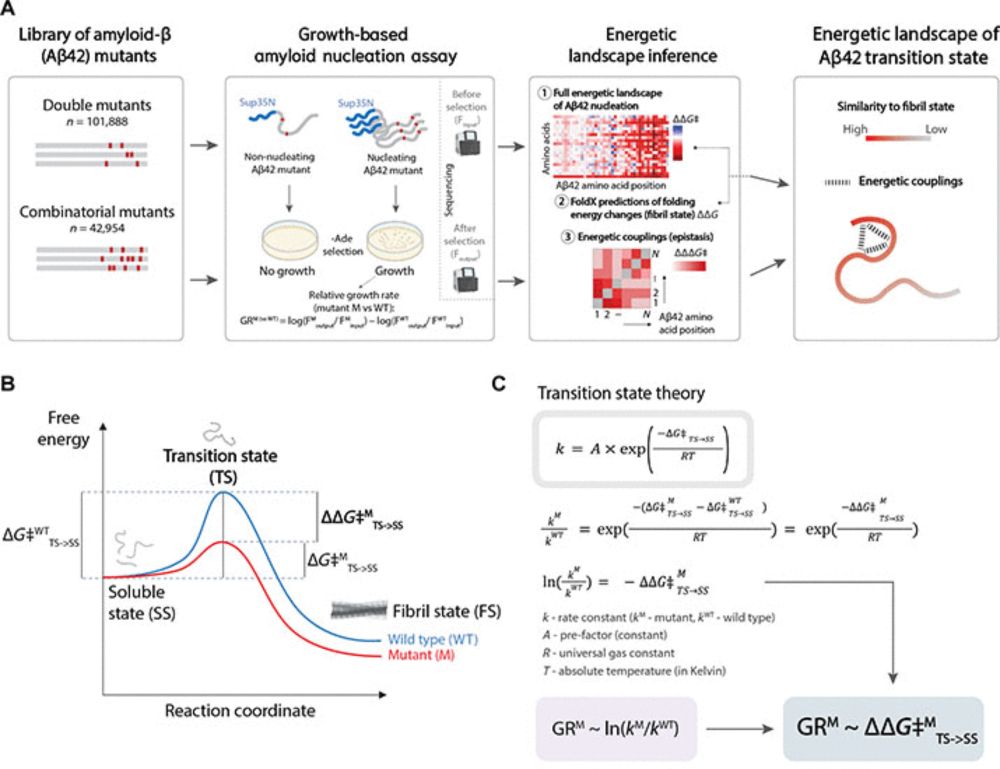
In our new study we cracked open the black box of early protein aggregation, and the findings could reshape how we fight neurodegeneration. 🧵👇
www.science.org/doi/10.1126/...
3,500 mutants, to single-site saturation 🧬
>45,000 binding and abundance measurements 📶
Very happy to present our latest work – where deep mutational scanning meets the world of small molecules.
www.biorxiv.org/content/10.1...
With @benlehner.bsky.social
[1/7]

3,500 mutants, to single-site saturation 🧬
>45,000 binding and abundance measurements 📶
Very happy to present our latest work – where deep mutational scanning meets the world of small molecules.
www.biorxiv.org/content/10.1...
With @benlehner.bsky.social
[1/7]
A new platform for high resolution functional mapping of GPCRs with massive mutagenesis. We identify the core activation network, residues involved in biased signaling, and generate >7,000 full dose response curves. With @benlehner.bsky.social

A new platform for high resolution functional mapping of GPCRs with massive mutagenesis. We identify the core activation network, residues involved in biased signaling, and generate >7,000 full dose response curves. With @benlehner.bsky.social

