
@SEBiolDev board member
I heard that getting in is one of the biggest challenges of all times in science 🤣
I heard that getting in is one of the biggest challenges of all times in science 🤣

👇👇

👇👇
So, we demonstrate that modulating protein stability can influence the expression of a downstream TF.

So, we demonstrate that modulating protein stability can influence the expression of a downstream TF.
Three day treatments with proteasomal inhibitors mostly killed the cells, BUT we observe signs of differentiation deceleration

Three day treatments with proteasomal inhibitors mostly killed the cells, BUT we observe signs of differentiation deceleration
✔️ differential abundance & stability of proteasomal particles
✔️ higher proteasomal activity in 🐭 compared to 👤 embryonic spinal cord
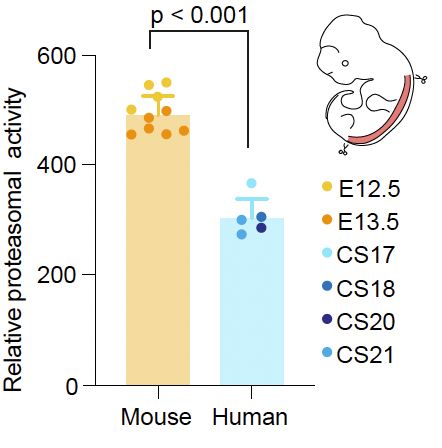
✔️ differential abundance & stability of proteasomal particles
✔️ higher proteasomal activity in 🐭 compared to 👤 embryonic spinal cord

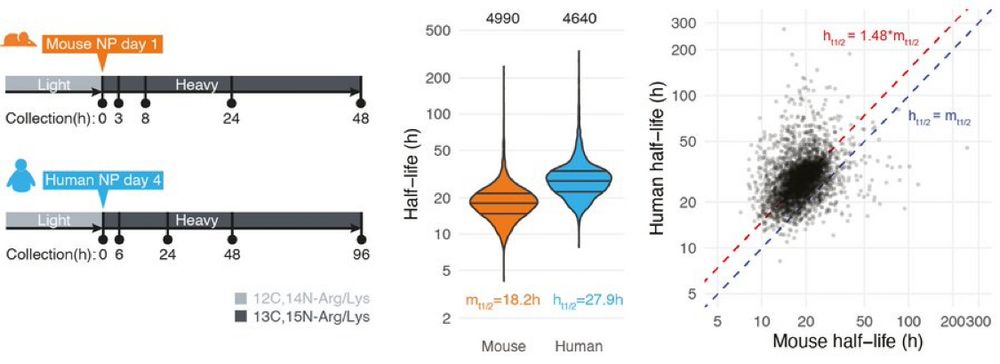

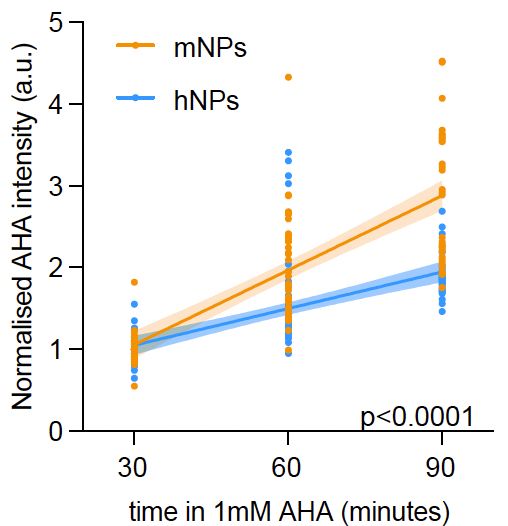
We find ~1.4-fold difference in decay and little differences in OLIG2 production. We also observed higher OLIG2 production rates in mouse.

We find ~1.4-fold difference in decay and little differences in OLIG2 production. We also observed higher OLIG2 production rates in mouse.


