
Chaotic good alignment, or so my lab tells me...

- Should be easily adapted to respond to other kinases.
- Should easily "plug into" other target proteins.
Both are true! For example, here is a "phospho-nanobody" that binds actin only when ERK phosphorylates it :)
- Should be easily adapted to respond to other kinases.
- Should easily "plug into" other target proteins.
Both are true! For example, here is a "phospho-nanobody" that binds actin only when ERK phosphorylates it :)
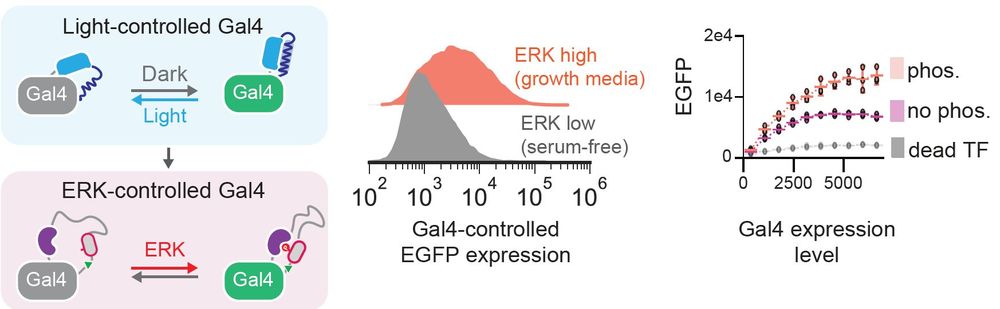
His final version basically looks as good as regular Gal4 when ERK is on, and has a 20x change in gene expression between ERK-on and off states!

His final version basically looks as good as regular Gal4 when ERK is on, and has a 20x change in gene expression between ERK-on and off states!
- Take an opto-Gal4 transcription factor we previously made by inserting the AsLOV2 switch
- Swap out AsLOV2 for a FRET biosensor for our favorite kinase, ERK
- Check if ERK activity now controls Gal4-induced gene expression!
It worked, but honestly, not that well...
- Take an opto-Gal4 transcription factor we previously made by inserting the AsLOV2 switch
- Swap out AsLOV2 for a FRET biosensor for our favorite kinase, ERK
- Check if ERK activity now controls Gal4-induced gene expression!
It worked, but honestly, not that well...

One way to do that is to fuse an "opto-switch" domain to a target protein. Light changes the conformation of the opto-switch, which tugs on the protein to turn it on or off!

One way to do that is to fuse an "opto-switch" domain to a target protein. Light changes the conformation of the opto-switch, which tugs on the protein to turn it on or off!
www.biorxiv.org/content/10.1...
We address a big challenge in synbio: If you give me a protein "X", how can I give you a version of X whose activity is controlled by a kinase?

www.biorxiv.org/content/10.1...
We address a big challenge in synbio: If you give me a protein "X", how can I give you a version of X whose activity is controlled by a kinase?


