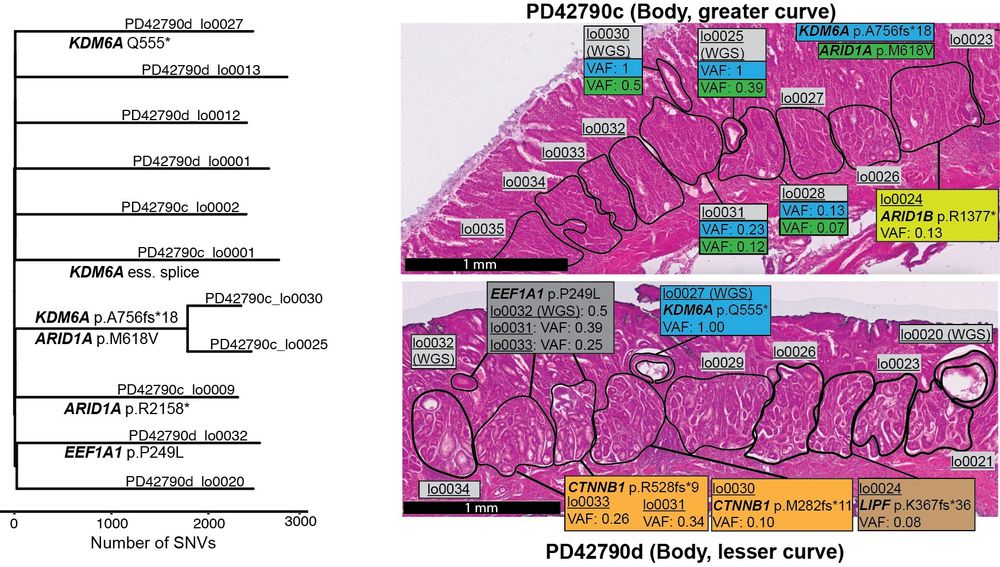
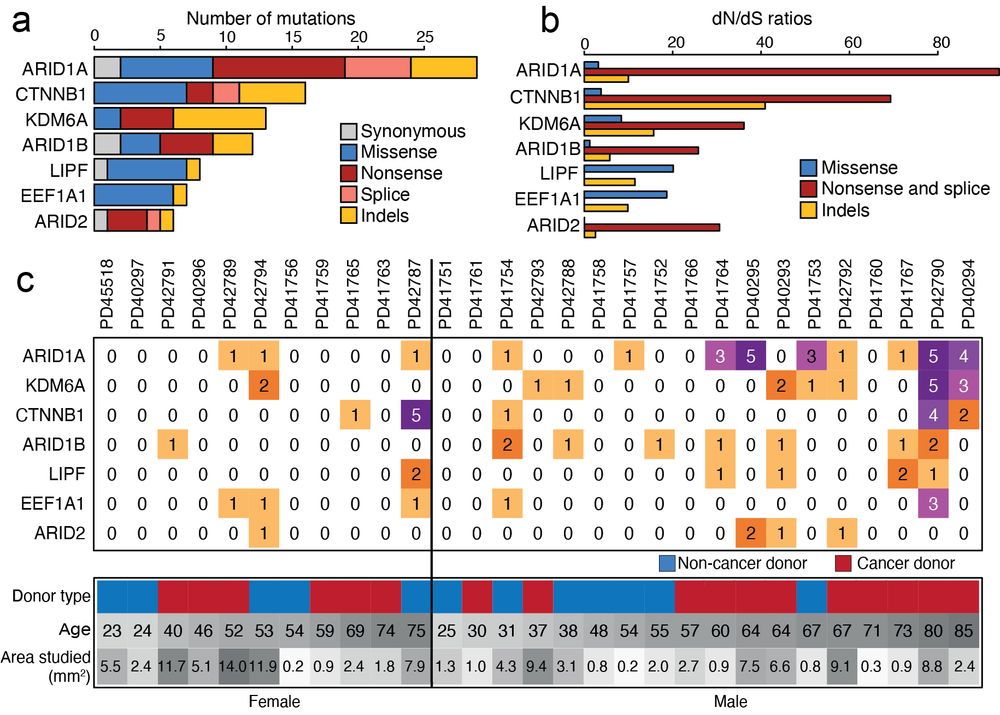
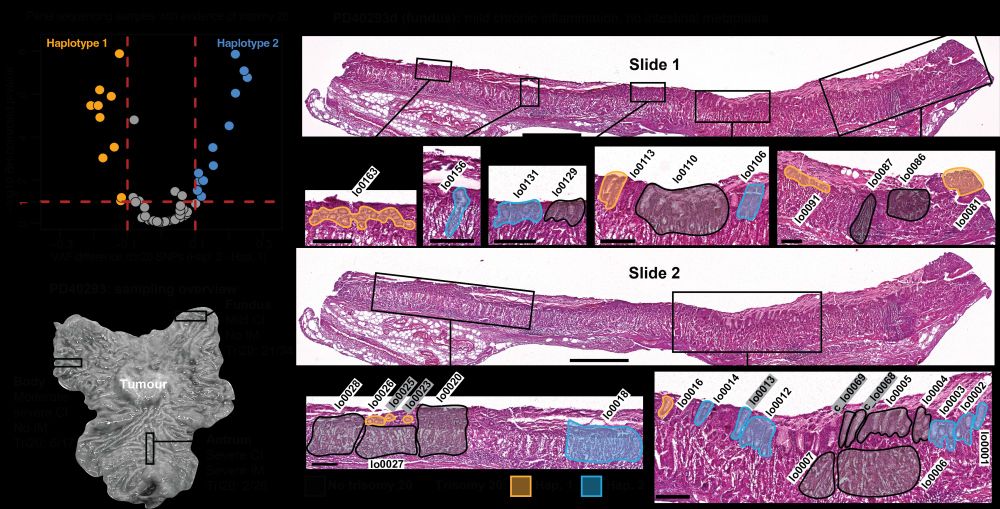
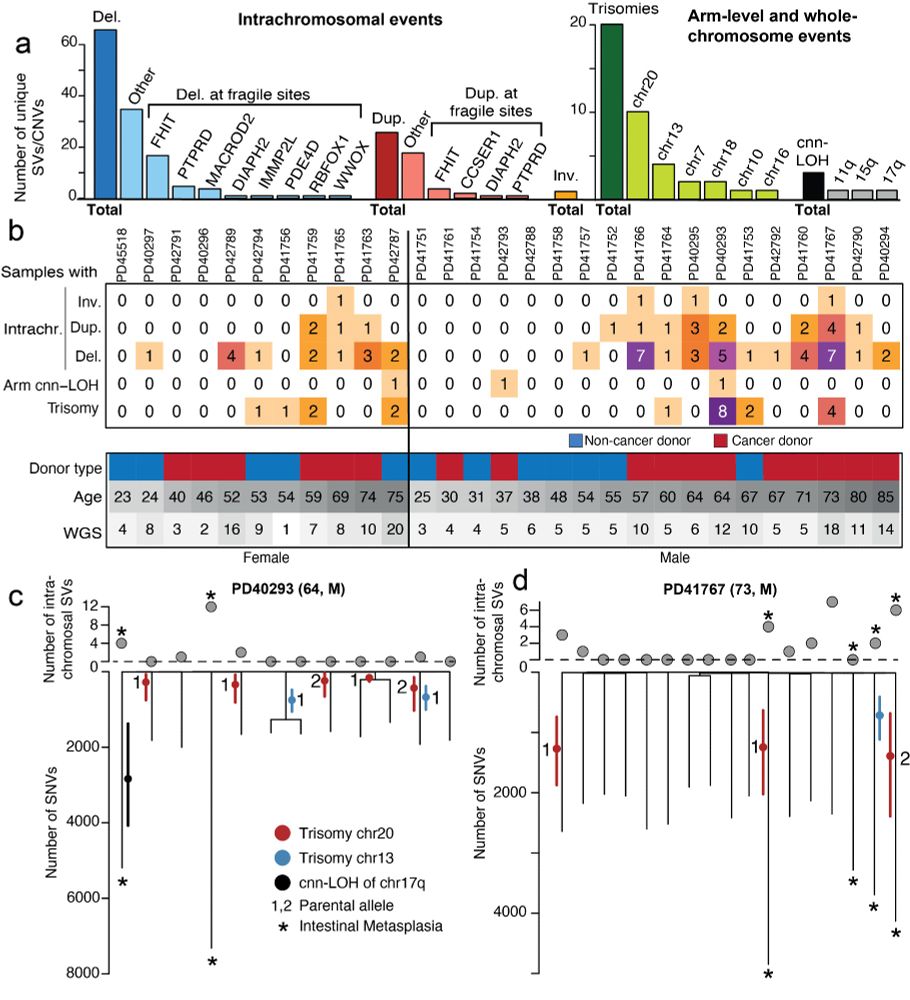
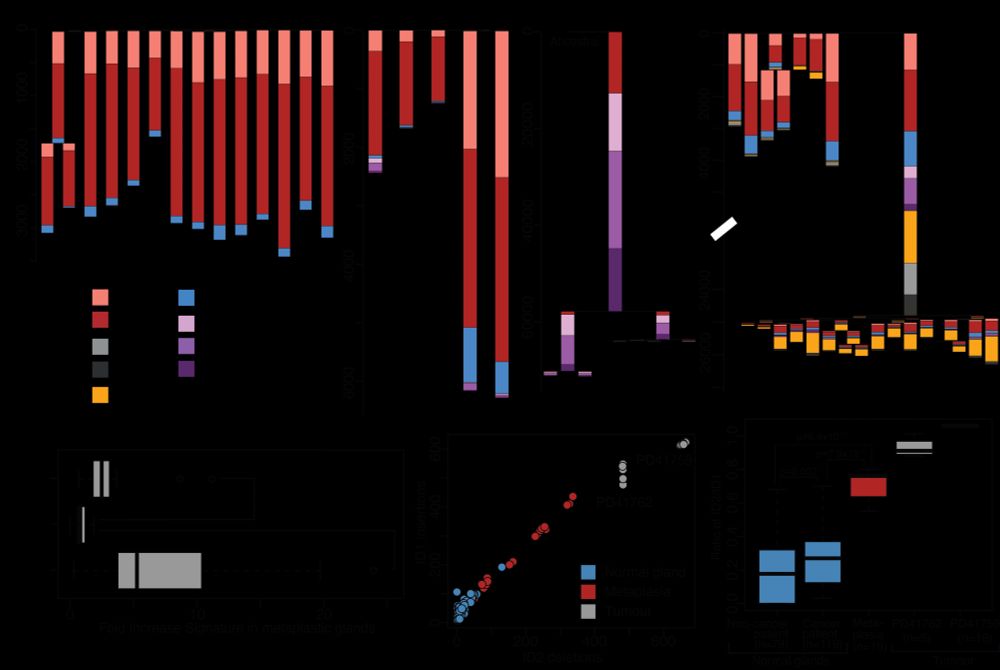
at Broad Institute | Former PhD student at Sanger Institute | Lineage tracing, cancer genomics, human development
@emblebi.bsky.social!
Reach out if interested or you have any questions, and apply here: embl.wd103.myworkdayjobs.com/en-US/EMBL/j...

@emblebi.bsky.social!
Reach out if interested or you have any questions, and apply here: embl.wd103.myworkdayjobs.com/en-US/EMBL/j...

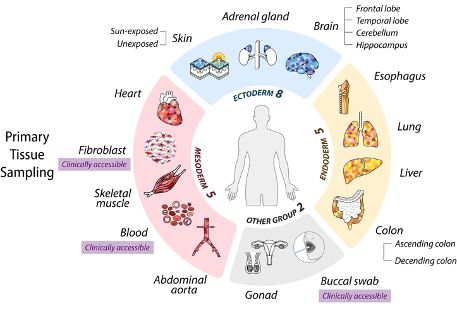
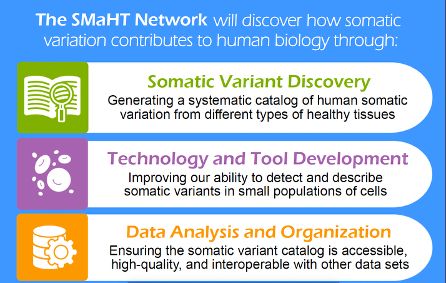
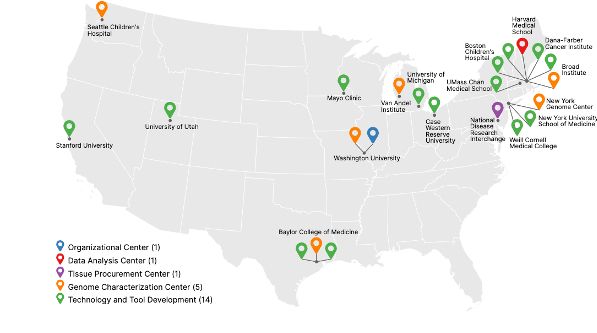
www.nature.com/articles/s41...
www.nature.com/articles/s41...