Metagenomics, gut microbiome, computational biology
Metalog: curated and harmonised contextual data for global metagenomics samples
now out in @narjournal.bsky.social
academic.oup.com/nar/advance-...

Metalog: curated and harmonised contextual data for global metagenomics samples
now out in @narjournal.bsky.social
academic.oup.com/nar/advance-...
In this study, we highlight that LDGs are not universal in marine microbiomes but reflect lineage-specific ecological strategies and environmental responses.
doi.org/10.1101/2025...

In this study, we highlight that LDGs are not universal in marine microbiomes but reflect lineage-specific ecological strategies and environmental responses.
doi.org/10.1101/2025...
#microbiome #microsky
doi.org/10.1128/msys...

#microbiome #microsky
doi.org/10.1128/msys...
Using ONT+Illumina, we get better MAGs than to corresponding species representative in public databases
doi.org/10.1101/2025...

Using ONT+Illumina, we get better MAGs than to corresponding species representative in public databases
doi.org/10.1101/2025...
#microsky
#microsky
www.biorxiv.org/content/10.1...
In this study, we present the largest systematic analysis of microbiome structure and function, integrating 85K uniformly processed metagenomes from diverse habitats worldwide.
@podlesny.bsky.social @jonas-bio.bsky.social @borklab.bsky.social

www.biorxiv.org/content/10.1...
In this study, we present the largest systematic analysis of microbiome structure and function, integrating 85K uniformly processed metagenomes from diverse habitats worldwide.
@podlesny.bsky.social @jonas-bio.bsky.social @borklab.bsky.social
Led by
@vishnuprasoodanan.bsky.social & @omaistrenko.bsky.social , we asked a question (almost) as old as microbiology: how many prokaryotic species exist on Earth? More specifically, how much diversity is "hiding" in existing metagenomic data?
A 🧵.
Led by
@vishnuprasoodanan.bsky.social & @omaistrenko.bsky.social , we asked a question (almost) as old as microbiology: how many prokaryotic species exist on Earth? More specifically, how much diversity is "hiding" in existing metagenomic data?
A 🧵.
🔬💊 Explore cutting-edge breakthroughs in #microbiome research, from methodological innovations to integrative approaches and personalised therapeutics. Join #EESMicrobiome 🦠🔍
➡️ https://s.embl.org/ees25-08-bl

🔬💊 Explore cutting-edge breakthroughs in #microbiome research, from methodological innovations to integrative approaches and personalised therapeutics. Join #EESMicrobiome 🦠🔍
➡️ https://s.embl.org/ees25-08-bl
rdcu.be/epoRR
@blekhman.bsky.social @sambhawa.bsky.social and Dr. Kelsey Johnson.
rdcu.be/epoRR
@blekhman.bsky.social @sambhawa.bsky.social and Dr. Kelsey Johnson.
📅 16 – 19 Sep 2025
📍 EMBL Heidelberg and Virtual
📥 Submit your abstract by 24 June
➡️ https://s.embl.org/ees25-08-bl

In our work, we discovered TANB77, a bacterial clade previously obscured by a polyphyletic grouping in conventional taxonomy, as a reliable biomarker across diverse immunotherapy recipient groups.
www.nature.com/articles/s41...
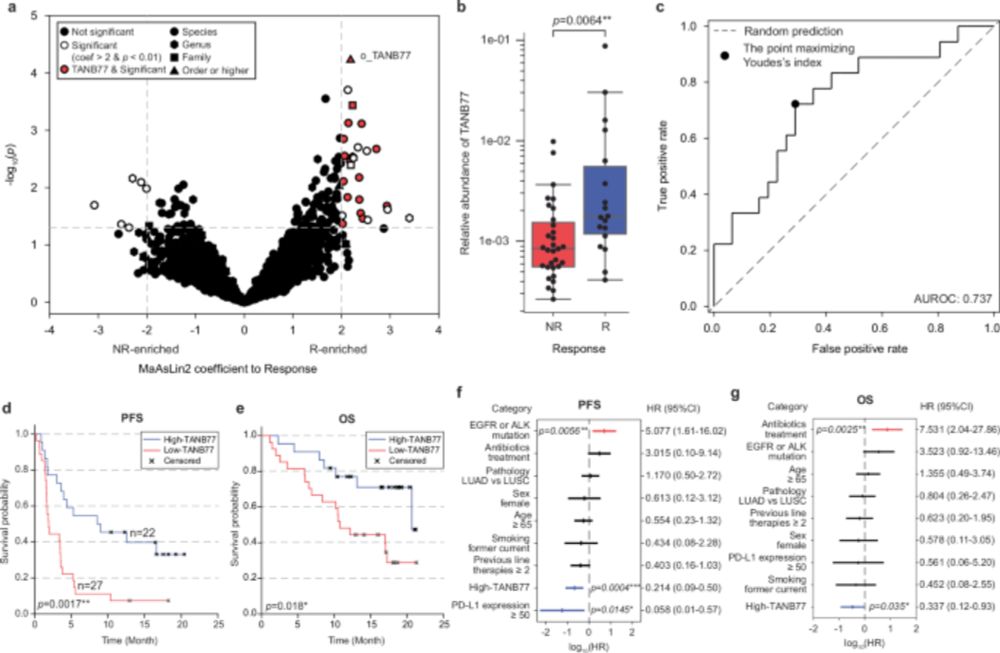
In our work, we discovered TANB77, a bacterial clade previously obscured by a polyphyletic grouping in conventional taxonomy, as a reliable biomarker across diverse immunotherapy recipient groups.
www.nature.com/articles/s41...

In the past two years, #EMBL scientists have gathered more than 3000 soil, sediment and water samples from Europe's coastlines within the #TREC expedition. (1/4)

In the past two years, #EMBL scientists have gathered more than 3000 soil, sediment and water samples from Europe's coastlines within the #TREC expedition. (1/4)
If you use microbiome data to look for disease associations, you might want to make sure microbial load is not confounding your results!
www.cell.com/cell/fulltex...
Congratulations @suguru-nishijima.bsky.social @borklab.bsky.social and others
#microsky #microbiome

If you use microbiome data to look for disease associations, you might want to make sure microbial load is not confounding your results!
www.cell.com/cell/fulltex...
Congratulations @suguru-nishijima.bsky.social @borklab.bsky.social and others
#microsky #microbiome
We developed a novel ML model that predicts fecal microbial load from relative species profiles, revealing it as a key factor shaping gut microbiome variation and disease-microbe associations
www.sciencedirect.com/science/arti...

We developed a novel ML model that predicts fecal microbial load from relative species profiles, revealing it as a key factor shaping gut microbiome variation and disease-microbe associations
www.sciencedirect.com/science/arti...
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

