
Kelly Chafin, Biosecurity Response
Chris Doering, Genomic Biosecurity
Siham Elhamoumi, Partnerships
Michael Gomez, Lab Technician
James Kremer, Laboratory Science
Kelly Chafin, Biosecurity Response
Chris Doering, Genomic Biosecurity
Siham Elhamoumi, Partnerships
Michael Gomez, Lab Technician
James Kremer, Laboratory Science


Earlier this year, we showed how our outward assembly pipeline can recover these genomes, testing the algorithm on a flagged SARS-CoV-2/plasmid construct where it did extremely well.

Earlier this year, we showed how our outward assembly pipeline can recover these genomes, testing the algorithm on a flagged SARS-CoV-2/plasmid construct where it did extremely well.

With four dedicated field samplers working most weekdays, we collect 300-800 nasal swabs weekly.
Viral reads are available via our dashboard: data.securebio.org/sampling-met...

With four dedicated field samplers working most weekdays, we collect 300-800 nasal swabs weekly.
Viral reads are available via our dashboard: data.securebio.org/sampling-met...
CASPER data identified measles in and West Nile Virus in two US communities:
health.hawaii.gov/news/newsroo...
content.govdelivery.com/accounts/MOD...
CASPER data identified measles in and West Nile Virus in two US communities:
health.hawaii.gov/news/newsroo...
content.govdelivery.com/accounts/MOD...
However, usability for time-sensitive investigations could be improved, which we’re planning to do this quarter.
Read more: naobservatory.org/blog/outward...
However, usability for time-sensitive investigations could be improved, which we’re planning to do this quarter.
Read more: naobservatory.org/blog/outward...
We identified the construct as a manufactured plasmid with a known genome.
Since we knew the genome we were supposed to recover, we had a rare chance to evaluate outward assembly in the real world.
We identified the construct as a manufactured plasmid with a known genome.
Since we knew the genome we were supposed to recover, we had a rare chance to evaluate outward assembly in the real world.
We will refine these estimates as we generate more sequencing data.
naobservatory.org/blog/swab-ba...
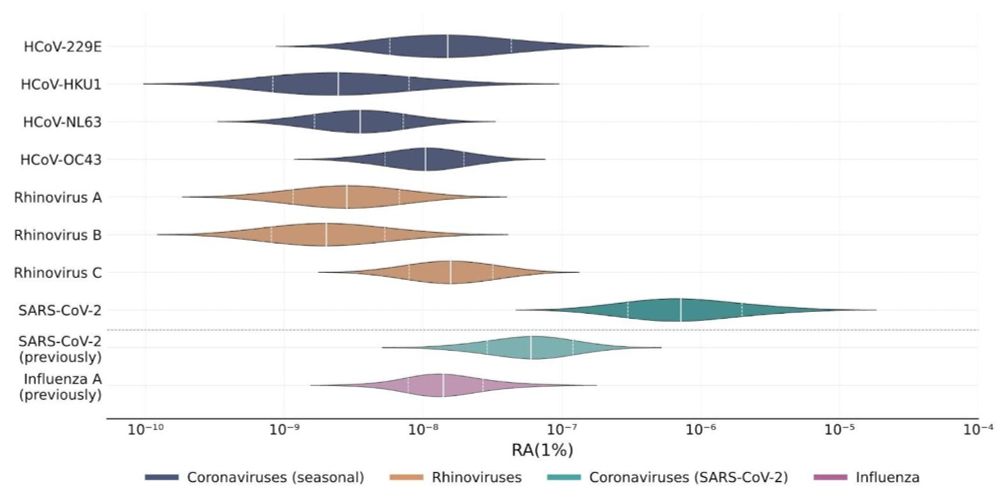
We will refine these estimates as we generate more sequencing data.
naobservatory.org/blog/swab-ba...
This has allowed us to collect 100-200 swabs per day. Over the coming quarter we will scale to additional weekdays and further optimize sampling.
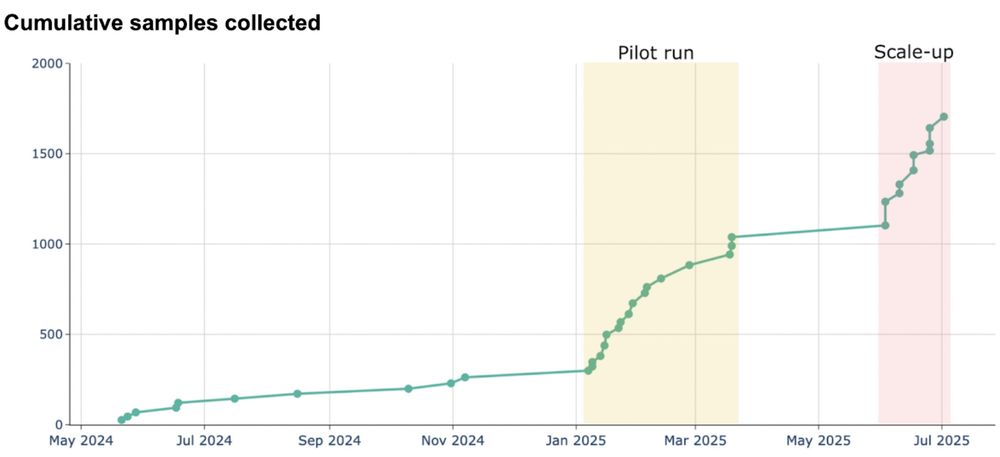
This has allowed us to collect 100-200 swabs per day. Over the coming quarter we will scale to additional weekdays and further optimize sampling.
Find the dashboard here: dholab.github.io/public_viz/
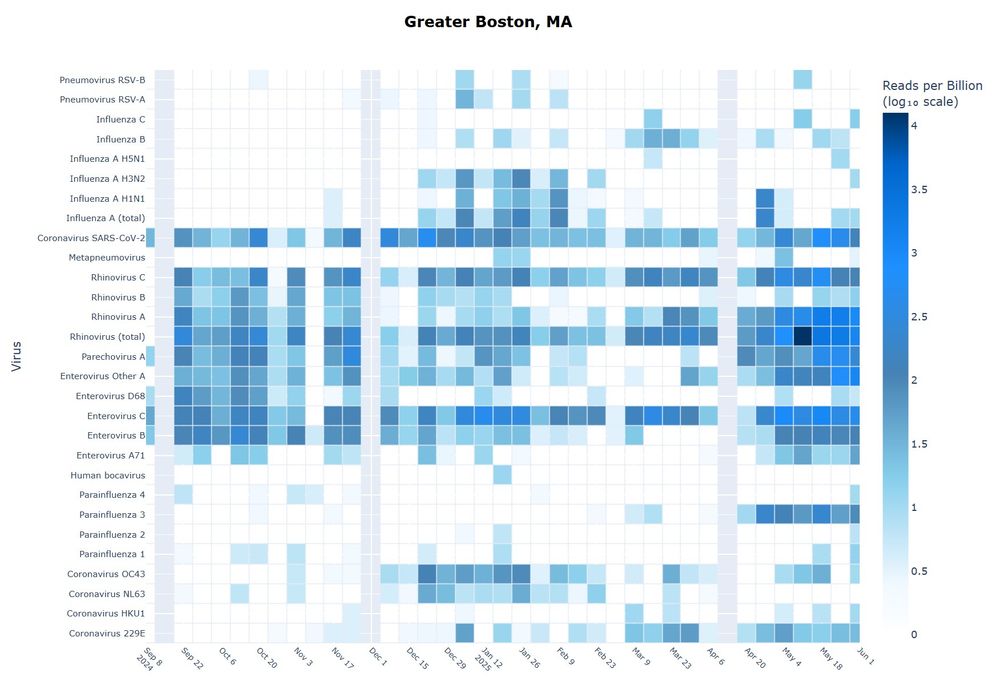
Find the dashboard here: dholab.github.io/public_viz/
With the new data, our genetic engineering detection system has now analyzed 892B read pairs, flagging 413 chimeras, with 43 “benign positives”.
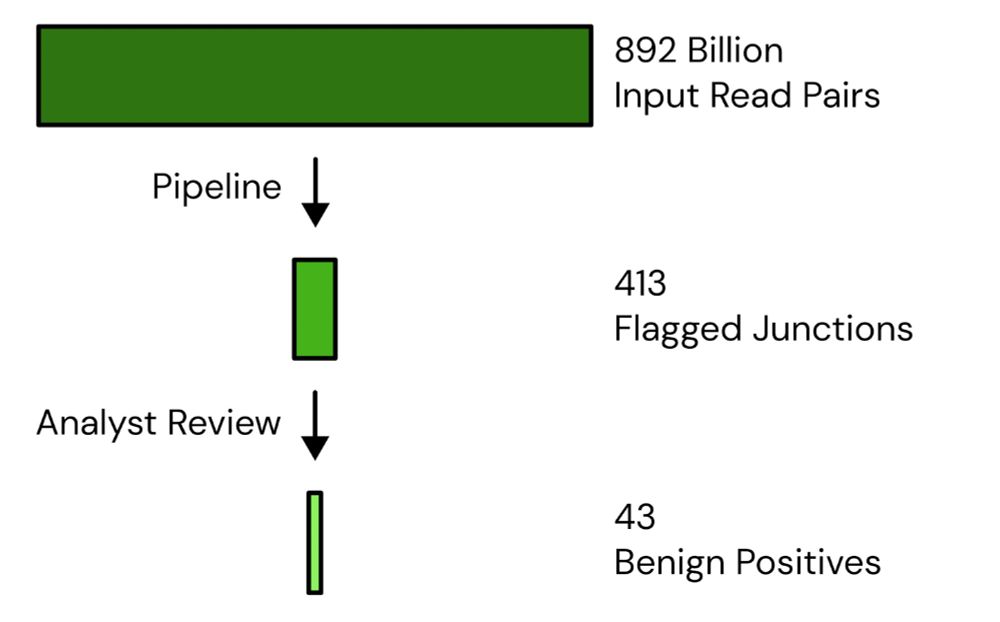
With the new data, our genetic engineering detection system has now analyzed 892B read pairs, flagging 413 chimeras, with 43 “benign positives”.
SARS-CoV-2 was again confirmed to be readily detectable, but cold viruses showed relatively low detectability.
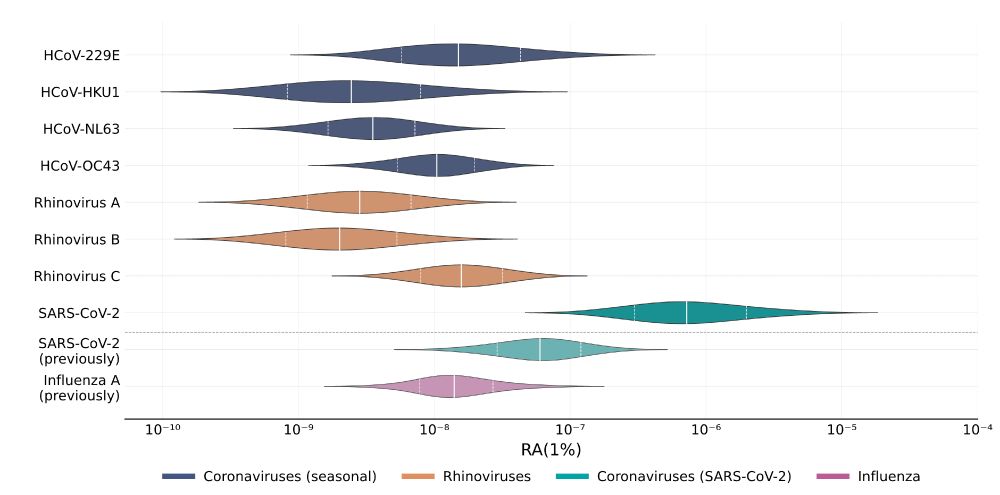
SARS-CoV-2 was again confirmed to be readily detectable, but cold viruses showed relatively low detectability.
We found that cold viruses are highly prevalent in winter: Many have a prevalence of 1% or higher!
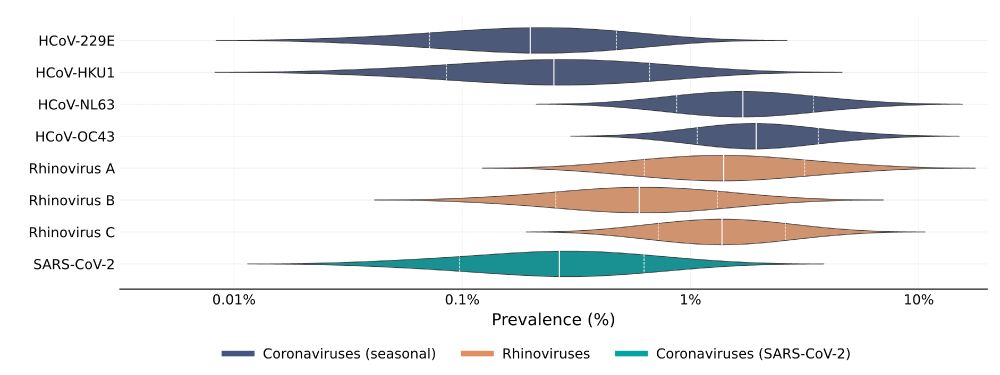
We found that cold viruses are highly prevalent in winter: Many have a prevalence of 1% or higher!
Collecting, pooling, and sequencing nasal swabs, we got a lot of information about the presence of cold viruses in the population, which we then linked with paired wastewater data.
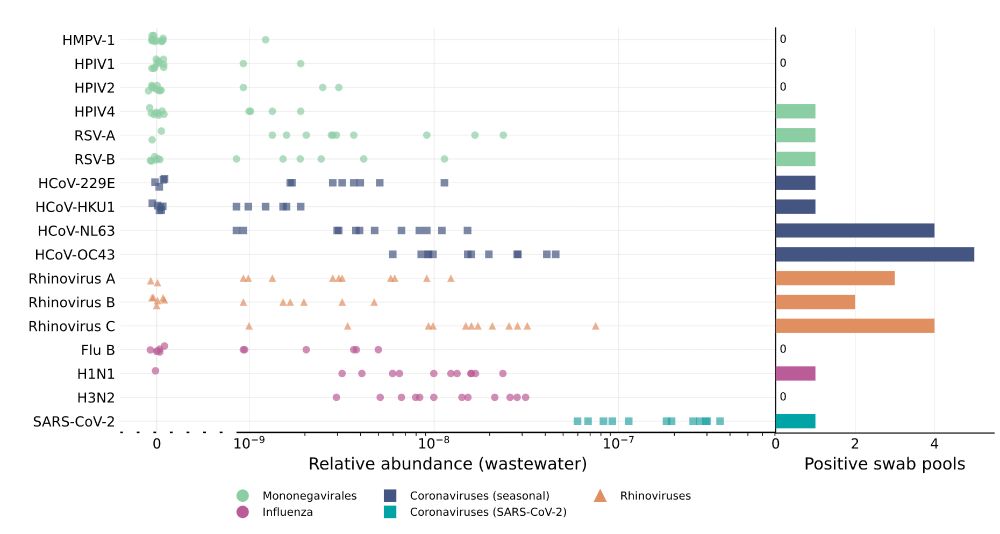
Collecting, pooling, and sequencing nasal swabs, we got a lot of information about the presence of cold viruses in the population, which we then linked with paired wastewater data.

