


Earlier this year, we showed how our outward assembly pipeline can recover these genomes, testing the algorithm on a flagged SARS-CoV-2/plasmid construct where it did extremely well.

Earlier this year, we showed how our outward assembly pipeline can recover these genomes, testing the algorithm on a flagged SARS-CoV-2/plasmid construct where it did extremely well.

With four dedicated field samplers working most weekdays, we collect 300-800 nasal swabs weekly.
Viral reads are available via our dashboard: data.securebio.org/sampling-met...

With four dedicated field samplers working most weekdays, we collect 300-800 nasal swabs weekly.
Viral reads are available via our dashboard: data.securebio.org/sampling-met...
- Major wastewater surveillance scaling: expanded to 31 sampling sites across 19 cities.
- Zephyr swab program scaling, now 400-800 swabs per week.
- New team members that lead response, wet-lab, and partnerships work.

- Major wastewater surveillance scaling: expanded to 31 sampling sites across 19 cities.
- Zephyr swab program scaling, now 400-800 swabs per week.
- New team members that lead response, wet-lab, and partnerships work.
We can recover these genomes with our outward assembly pipeline.
We recently tested outward assembly on a flagged SARS-CoV-2/plasmid construct – it did extremely well!
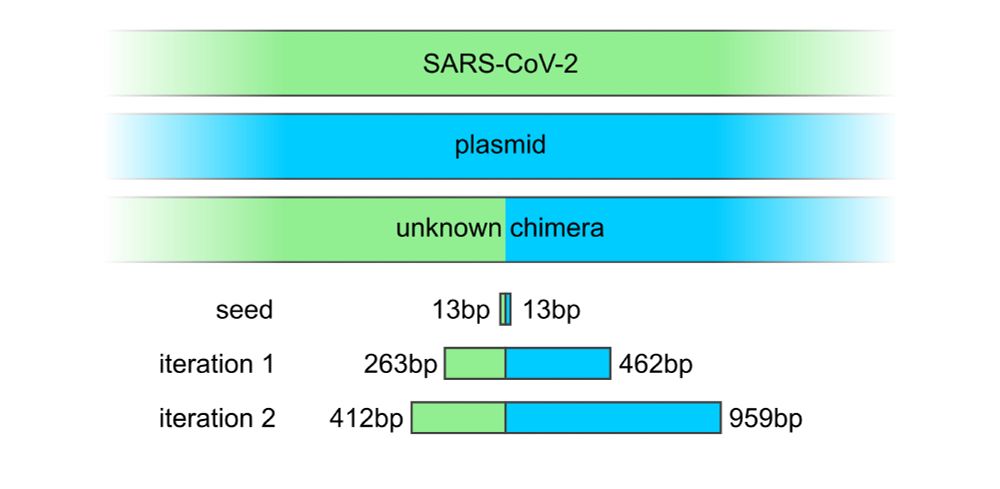
We can recover these genomes with our outward assembly pipeline.
We recently tested outward assembly on a flagged SARS-CoV-2/plasmid construct – it did extremely well!
We will refine these estimates as we generate more sequencing data.
naobservatory.org/blog/swab-ba...
We will refine these estimates as we generate more sequencing data.
naobservatory.org/blog/swab-ba...
This has allowed us to collect 100-200 swabs per day. Over the coming quarter we will scale to additional weekdays and further optimize sampling.
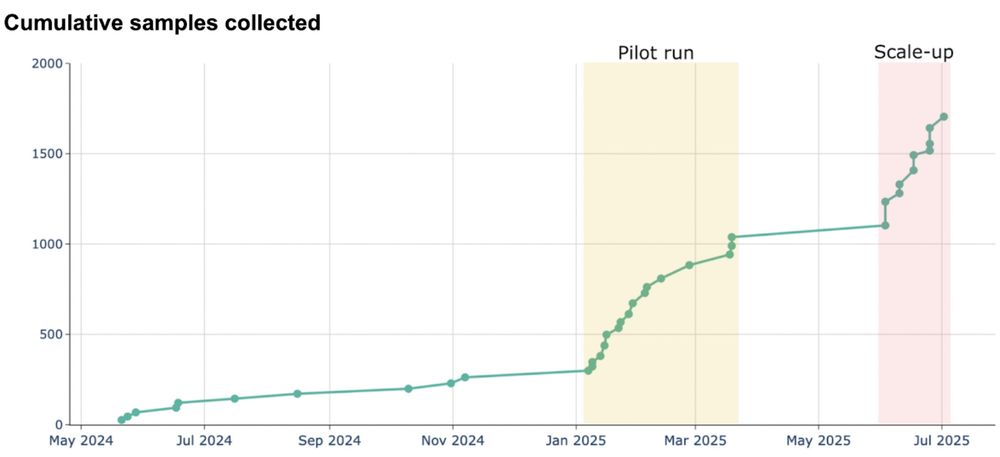
This has allowed us to collect 100-200 swabs per day. Over the coming quarter we will scale to additional weekdays and further optimize sampling.
Find the dashboard here: dholab.github.io/public_viz/
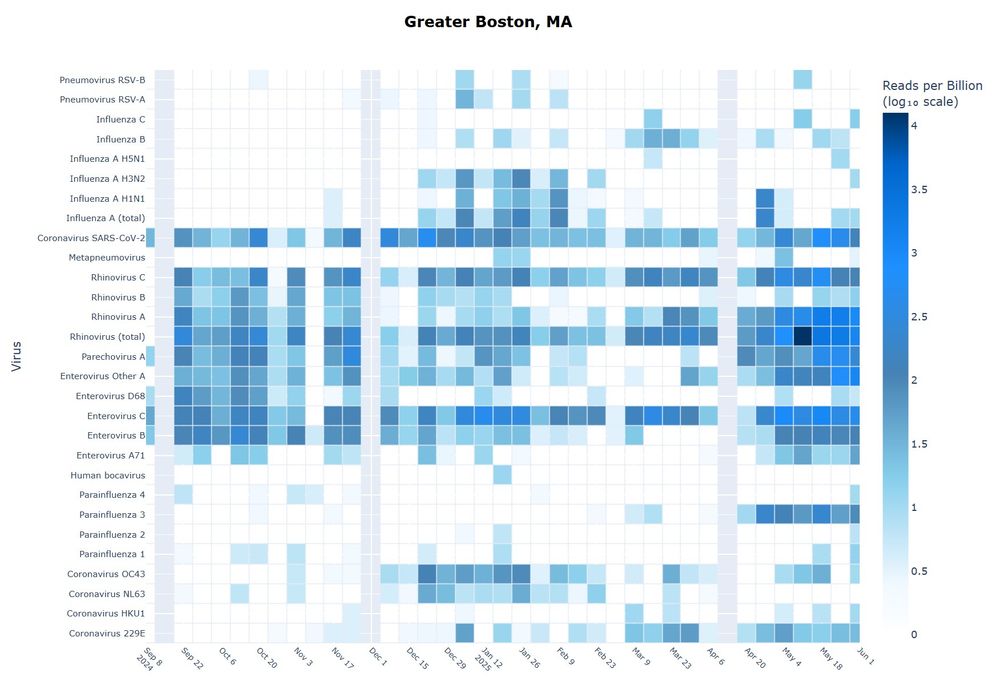
Find the dashboard here: dholab.github.io/public_viz/
With the new data, our genetic engineering detection system has now analyzed 892B read pairs, flagging 413 chimeras, with 43 “benign positives”.
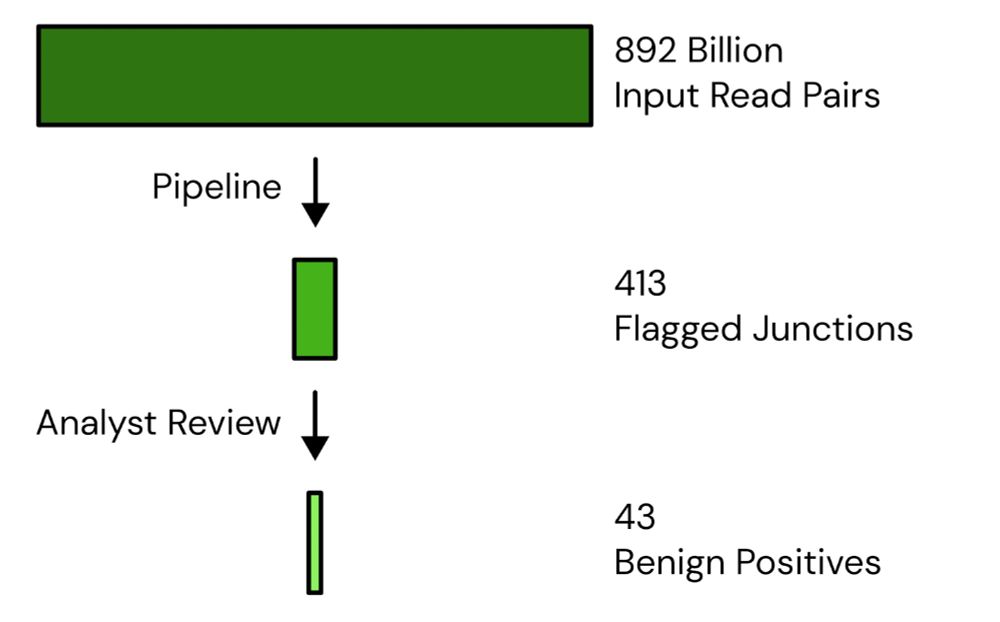
With the new data, our genetic engineering detection system has now analyzed 892B read pairs, flagging 413 chimeras, with 43 “benign positives”.
- We’ve further increased our sequencing capacity, producing 487B read pairs.
- Our partners created dashboards that summarize which pathogens we routinely see in wastewater.
- We’ve been scaling up our Boston-based swab sampling program.

- We’ve further increased our sequencing capacity, producing 487B read pairs.
- Our partners created dashboards that summarize which pathogens we routinely see in wastewater.
- We’ve been scaling up our Boston-based swab sampling program.
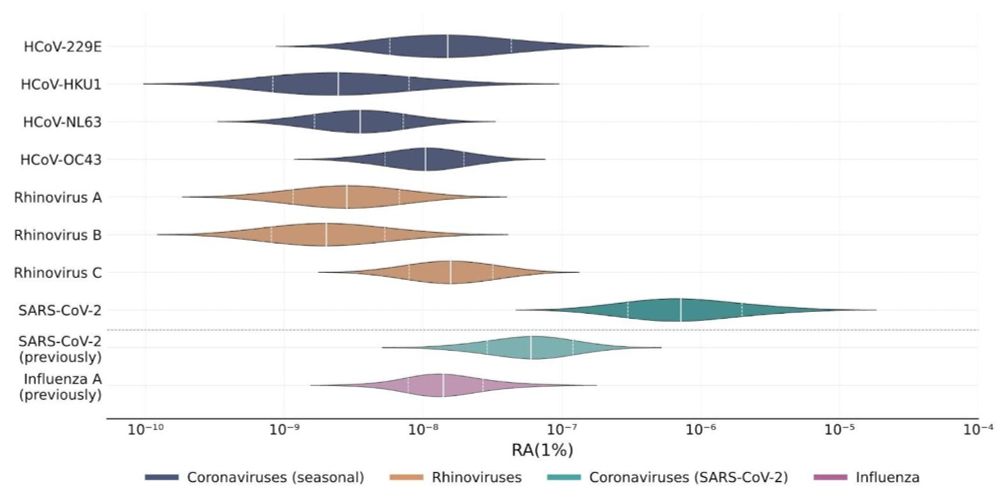
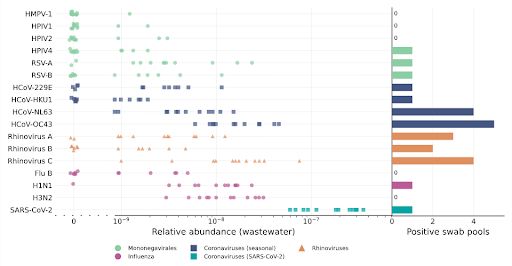
SARS-CoV-2 was again confirmed to be readily detectable, but cold viruses showed relatively low detectability.
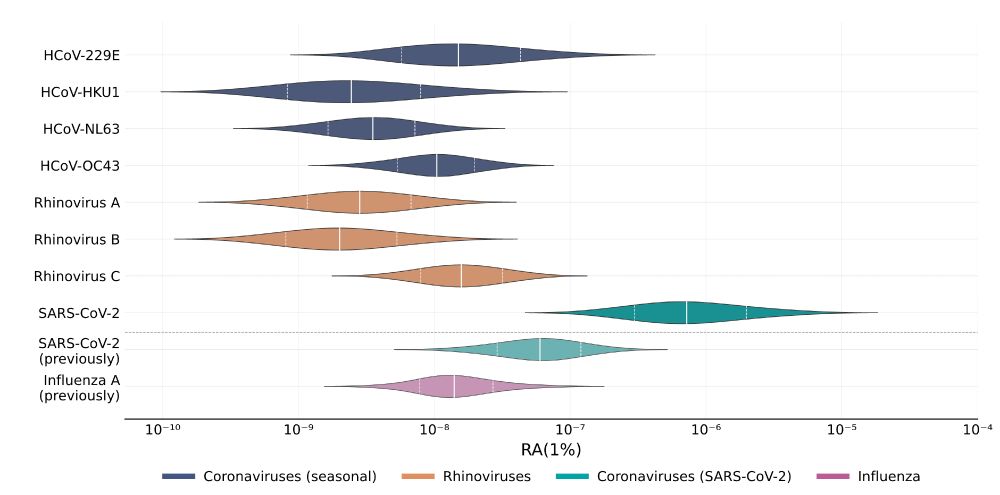
SARS-CoV-2 was again confirmed to be readily detectable, but cold viruses showed relatively low detectability.
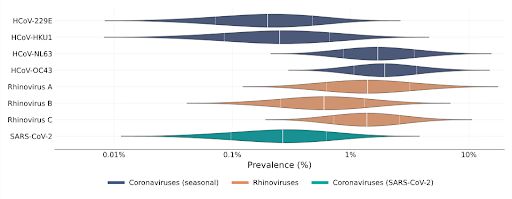
We found that cold viruses are highly prevalent in winter: Many have a prevalence of 1% or higher!
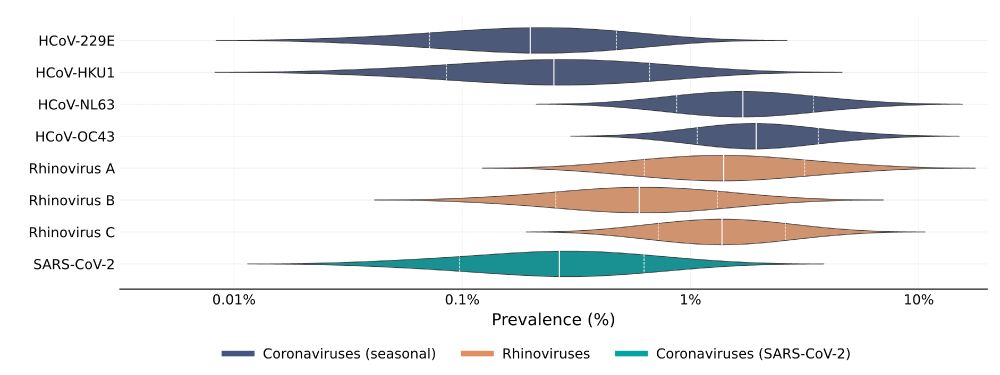
We found that cold viruses are highly prevalent in winter: Many have a prevalence of 1% or higher!
Collecting, pooling, and sequencing nasal swabs, we got a lot of information about the presence of cold viruses in the population, which we then linked with paired wastewater data.
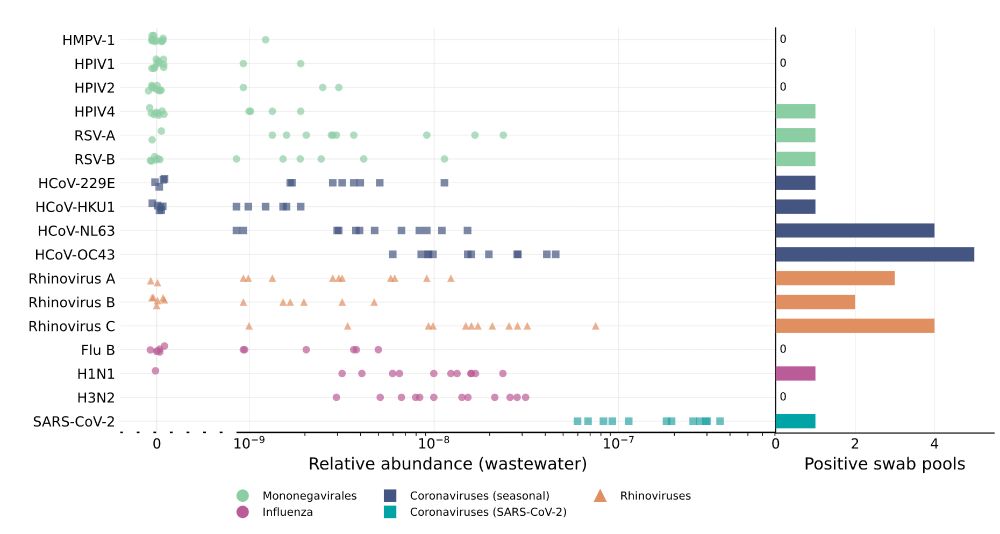
Collecting, pooling, and sequencing nasal swabs, we got a lot of information about the presence of cold viruses in the population, which we then linked with paired wastewater data.
SARS-CoV-2 is again highly detectable, but common cold viruses are harder to detect.
We will use this research to compare wastewater sequencing to other detection strategies.

SARS-CoV-2 is again highly detectable, but common cold viruses are harder to detect.
We will use this research to compare wastewater sequencing to other detection strategies.

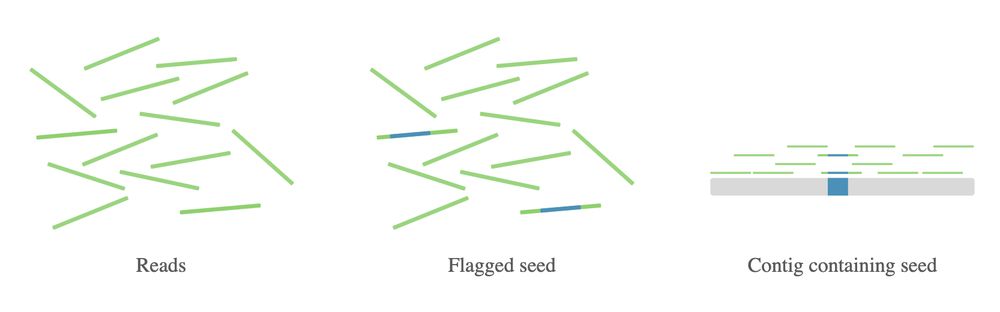
Instead, our approach searches billions of reads to find only those relevant to the seed, then assembles just those reads. 3/4
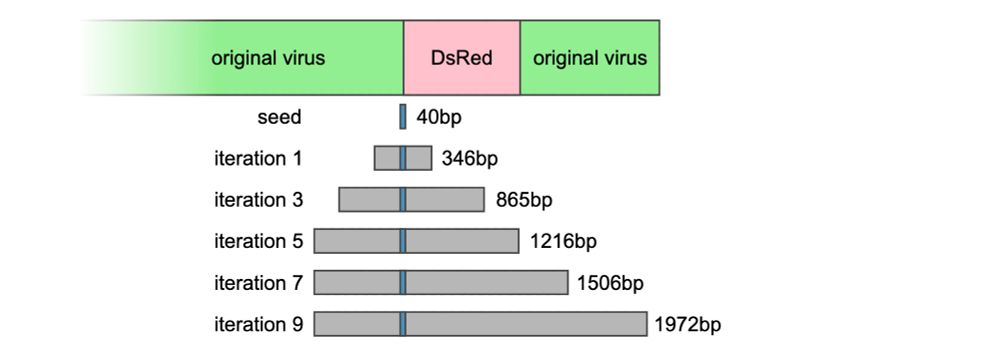
Instead, our approach searches billions of reads to find only those relevant to the seed, then assembles just those reads. 3/4

We built a new benchmark to answer that question.
To our surprise, we found that leading models outperform the vast majority of practicing virologists we sampled. 🧵 1/13

We built a new benchmark to answer that question.
To our surprise, we found that leading models outperform the vast majority of practicing virologists we sampled. 🧵 1/13

