www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...


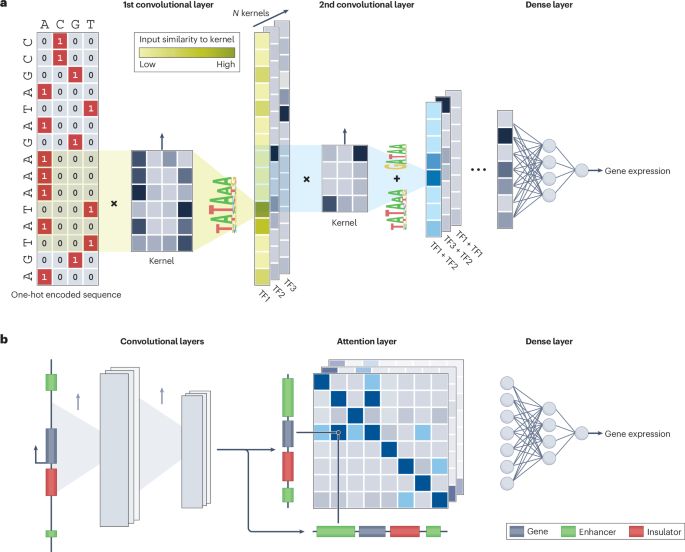

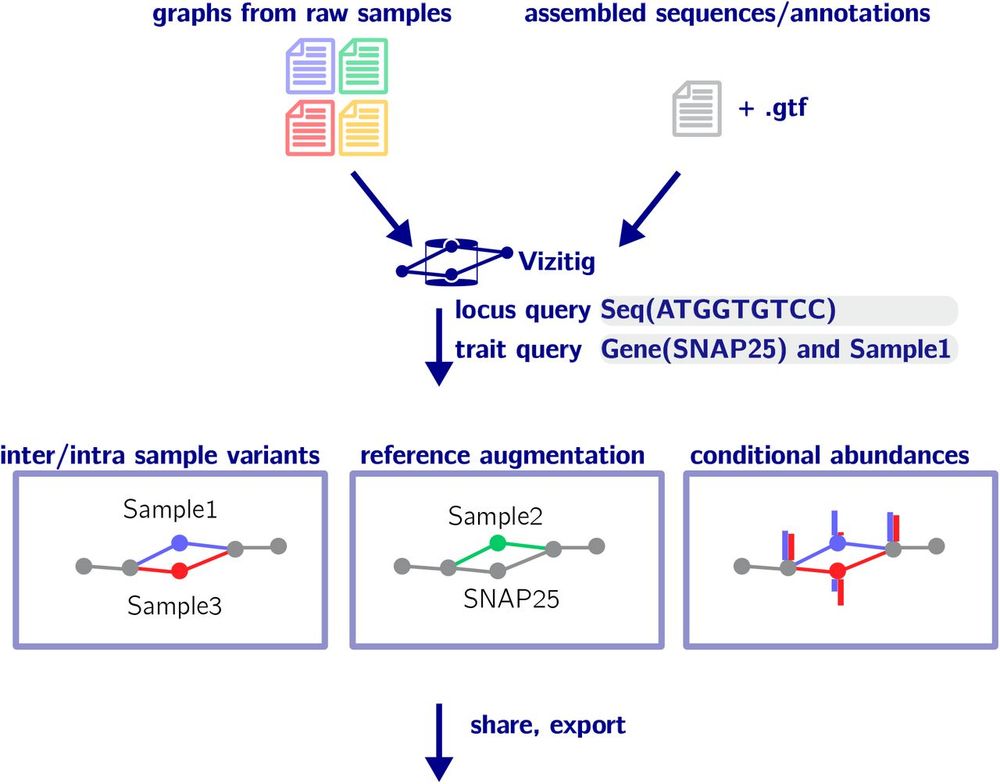
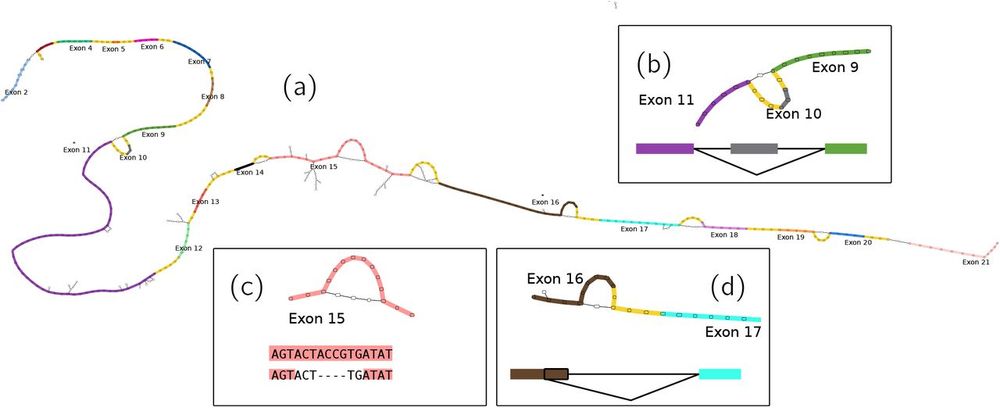
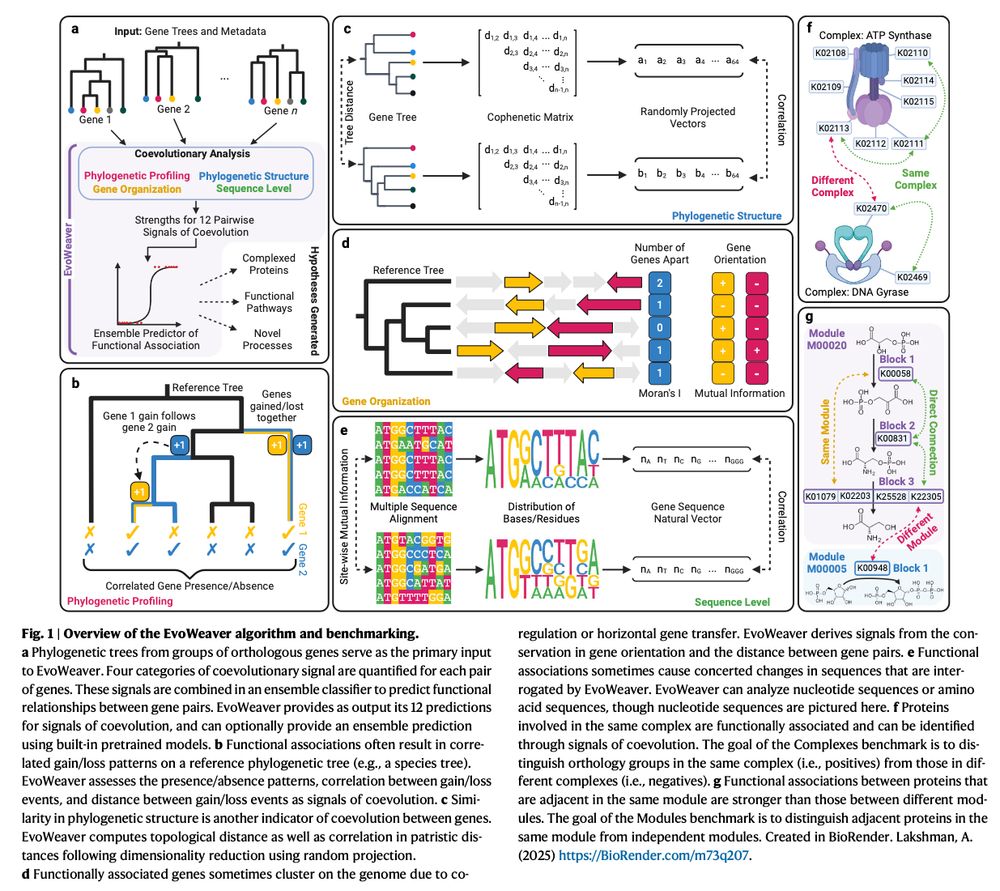







www.nature.com/articles/s41...

www.nature.com/articles/s41...






Branching architecture limits the rate of somatic mutation accumulation in trees
doi.org/10.1101/2025...
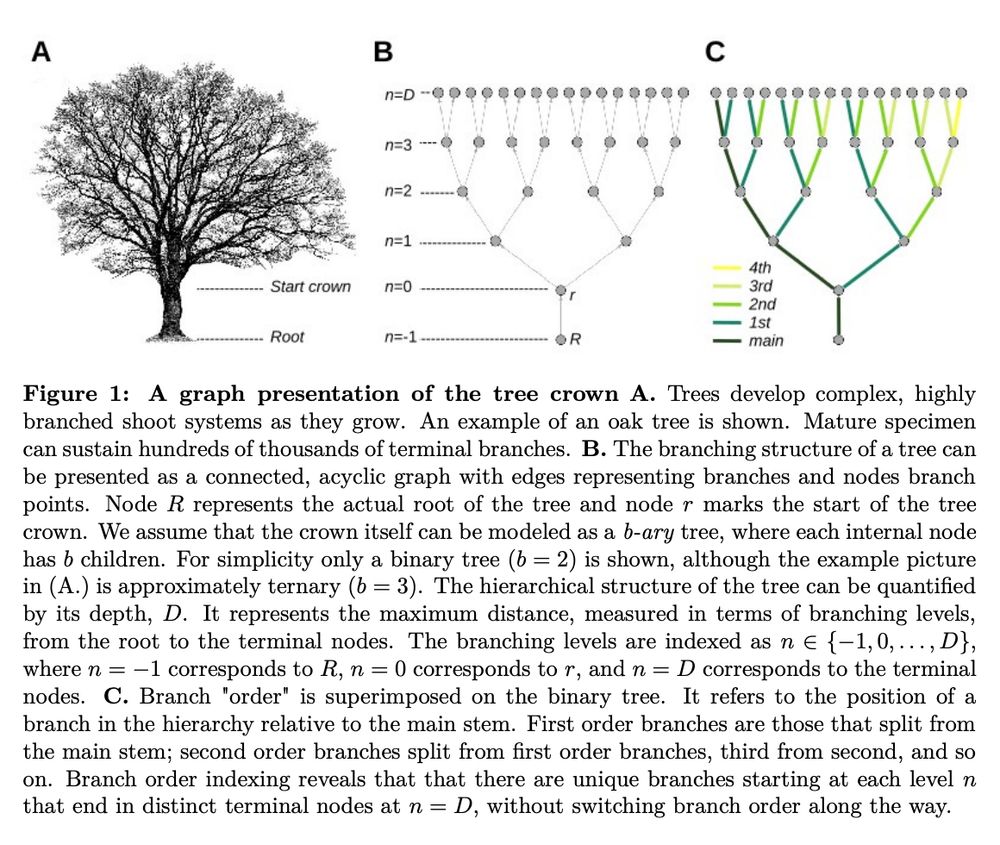
Branching architecture limits the rate of somatic mutation accumulation in trees
doi.org/10.1101/2025...





