PI at Boston Children’s Hospital SCP
Assistant Professor at Harvard SCRB












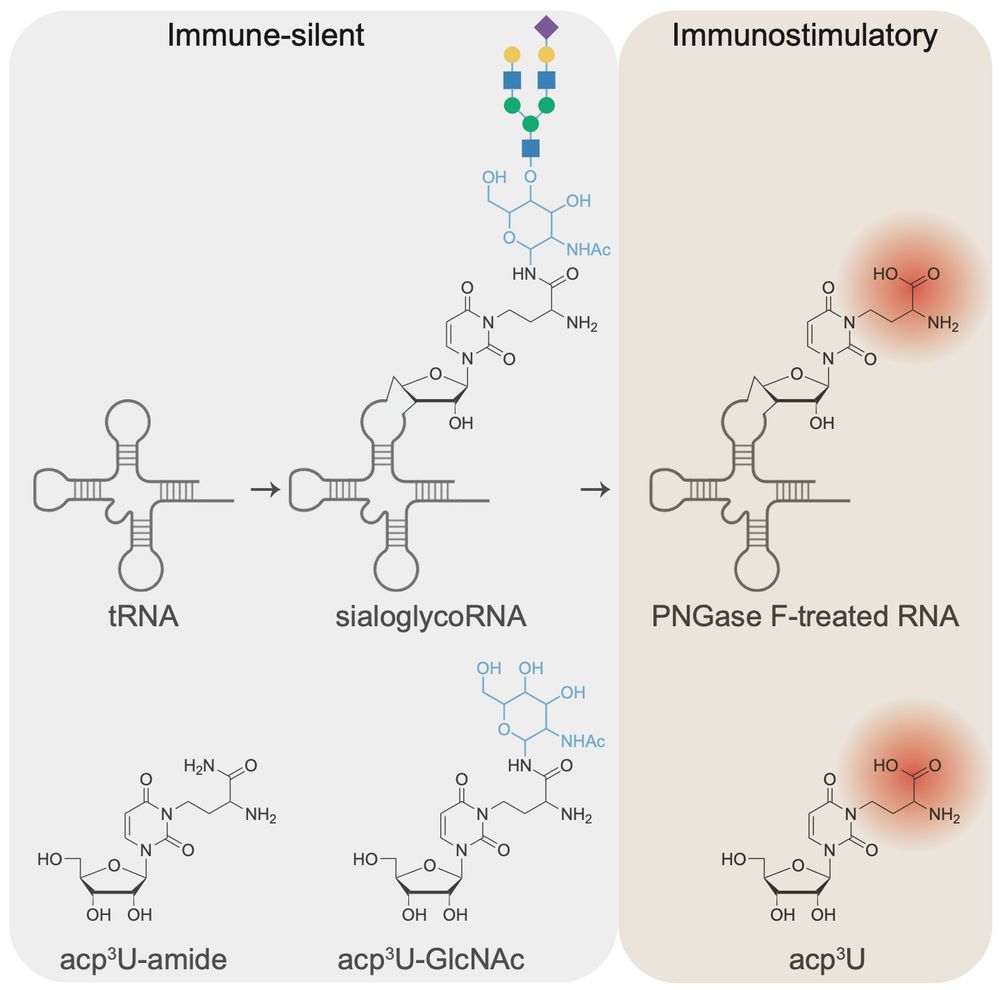


connecting ‘cell surface RNA biology’ to cancer biology
@natbiotech.nature.com
www.nature.com/articles/s41...
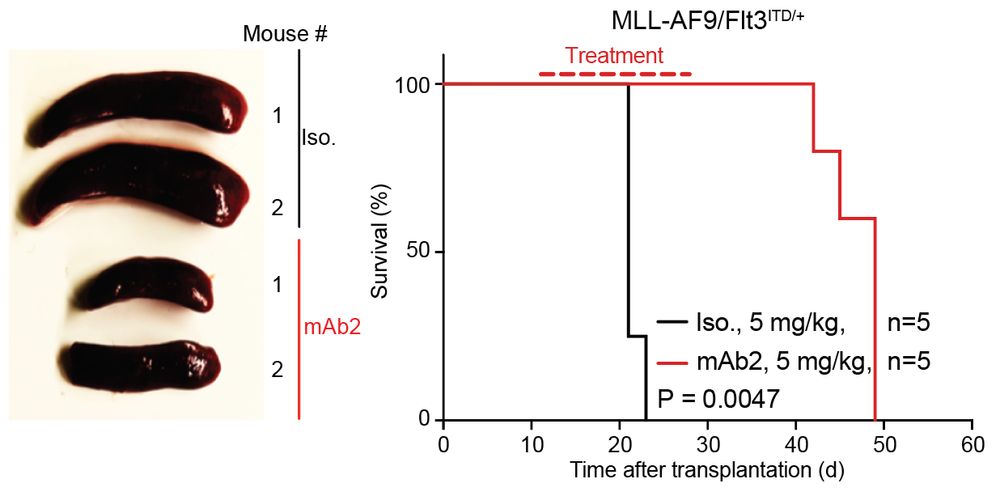
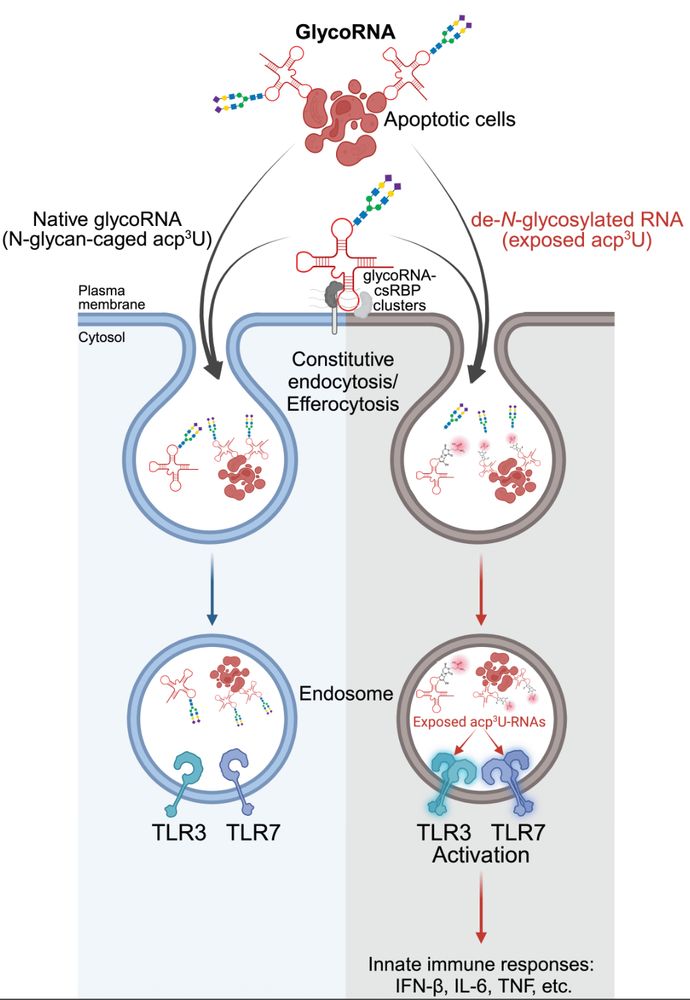
connecting ‘cell surface RNA biology’ to cancer biology
@natbiotech.nature.com
www.nature.com/articles/s41...
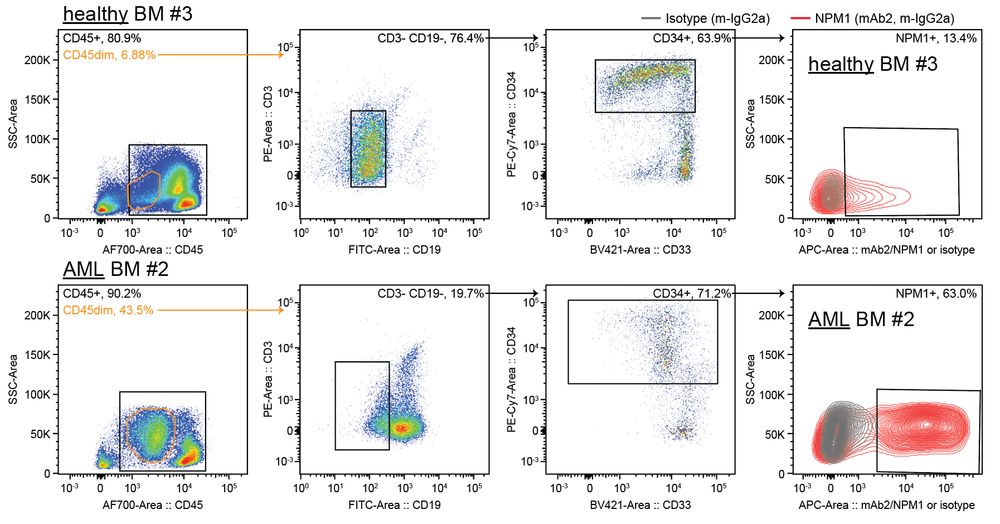
Enables ‘watching’ of RNP assemblies inside living cells
Special congrats to Luca Ducoli, Brian Zarnegar, and Paul Khavari @nature.com
www.nature.com/articles/s41...
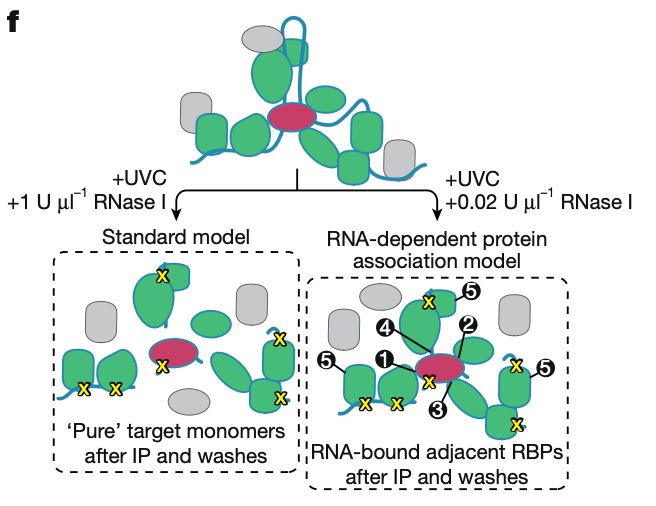
Enables ‘watching’ of RNP assemblies inside living cells
Special congrats to Luca Ducoli, Brian Zarnegar, and Paul Khavari @nature.com
www.nature.com/articles/s41...

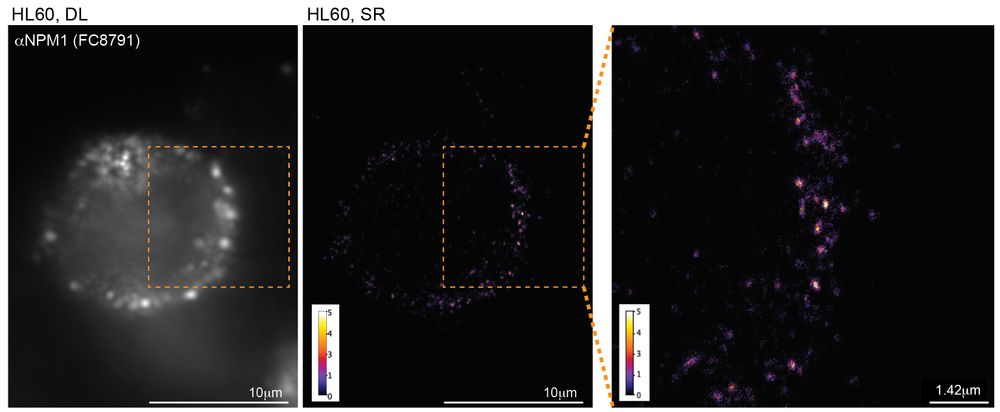
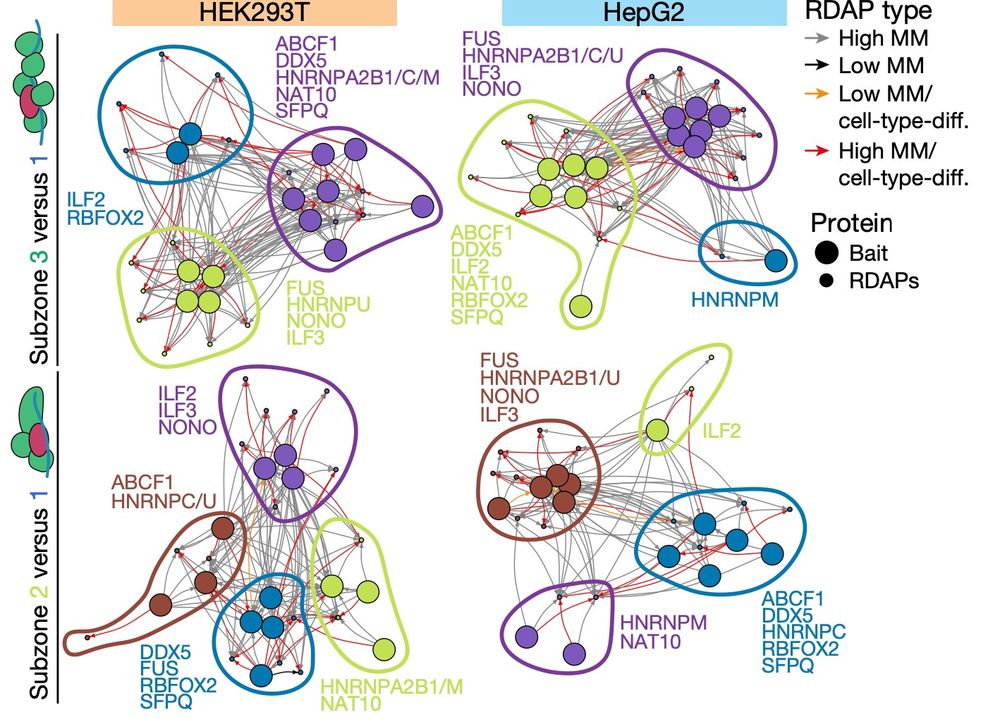
High resolution, High Effort, and Creative Analytics produce new views of the cell surface
www.biorxiv.org/content/10.1...

High resolution, High Effort, and Creative Analytics produce new views of the cell surface
www.biorxiv.org/content/10.1...








