
Metagenomics, protein evolution, ribosomes, and recently microbiome and its role in health.
https://orcid.org/0000-0002-9027-3824
Check it out: doi.org/10.1093/isme...

Check it out: doi.org/10.1093/isme...
Huge thanks to @ppenev.bsky.social @Amos Nissley @Dipti Nayak @Rohan Sachdeva @jhdcate.bsky.social @Jill Banfield @banfieldlab.bsky.social !!

Huge thanks to @ppenev.bsky.social @Amos Nissley @Dipti Nayak @Rohan Sachdeva @jhdcate.bsky.social @Jill Banfield @banfieldlab.bsky.social !!

rdcu.be/d5put
A thread…
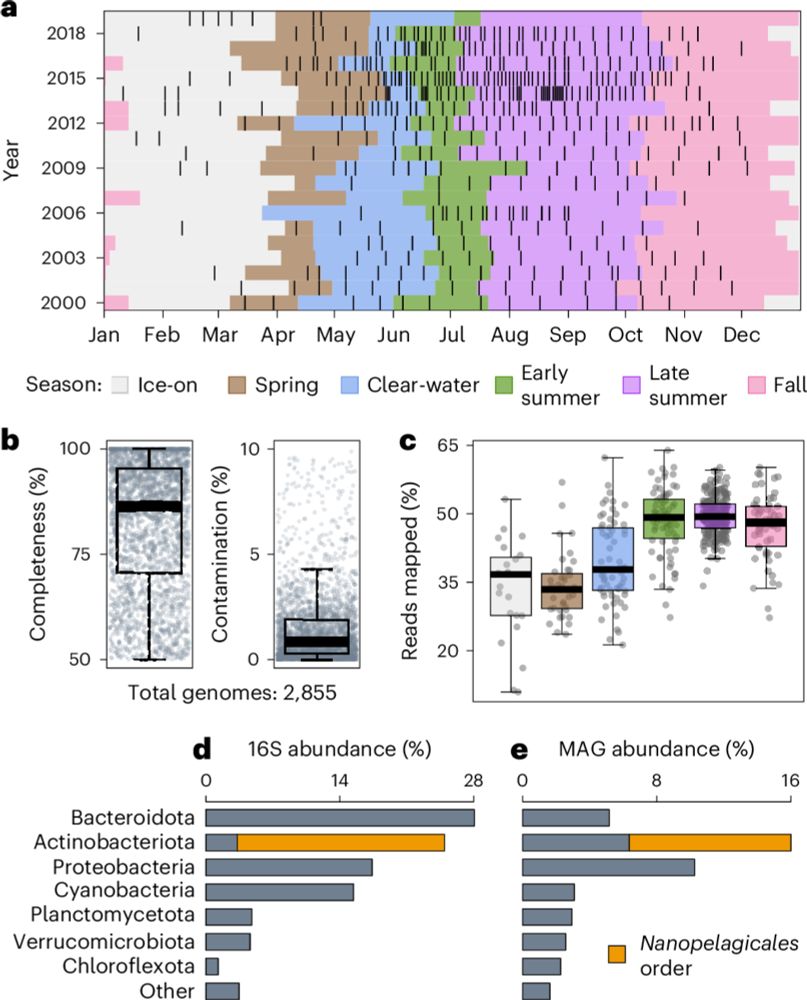
rdcu.be/d5put
A thread…
2 bioinfo PhD positions @ProbstLab @RESIST_CRC
#1 on microbial & epigenetic stress response in rivers
tinyurl.com/bdf8eu5a
#2 on virus-host stress response in rivers
tinyurl.com/43cjd6es (supervision Moraru)
Please share!
2 bioinfo PhD positions @ProbstLab @RESIST_CRC
#1 on microbial & epigenetic stress response in rivers
tinyurl.com/bdf8eu5a
#2 on virus-host stress response in rivers
tinyurl.com/43cjd6es (supervision Moraru)
Please share!

RNA language models predict mutations that improve RNA function
www.nature.com/articles/s41...
Congrats to @kshulgina.bsky.social @jhdcate.bsky.social and coauthors on a great C-GEM + @innovativegenomics.bsky.social collaboration! #NSFfunded #RNAsky

RNA language models predict mutations that improve RNA function
www.nature.com/articles/s41...
Congrats to @kshulgina.bsky.social @jhdcate.bsky.social and coauthors on a great C-GEM + @innovativegenomics.bsky.social collaboration! #NSFfunded #RNAsky
https://www.nature.com/articles/nrmicro.2017.133

https://www.nature.com/articles/nrmicro.2017.133
Celebrating the final ‘cosm harvest from our 7th/last 13CO2 experiment of 2024. We’re studying how the psY gene affects CH4 fluxes and soil C cycles.
Much gratitude to Rina Estera, Flor Ercoli, Pam Ronald Lab, Dave Savage Lab & many others @innovativegenomics.bsky.social




Celebrating the final ‘cosm harvest from our 7th/last 13CO2 experiment of 2024. We’re studying how the psY gene affects CH4 fluxes and soil C cycles.
Much gratitude to Rina Estera, Flor Ercoli, Pam Ronald Lab, Dave Savage Lab & many others @innovativegenomics.bsky.social


onlinelibrary.wiley.com/doi/full/10....

onlinelibrary.wiley.com/doi/full/10....

- Autotrophic biofilms sustained by deeply sourced groundwater host diverse bacteria implicated in sulfur and hydrogen metabolism
microbiomejournal.biomedcentral.com/articles/10....

- Autotrophic biofilms sustained by deeply sourced groundwater host diverse bacteria implicated in sulfur and hydrogen metabolism
microbiomejournal.biomedcentral.com/articles/10....
www.cell.com/iscience/ful...
www.cell.com/iscience/ful...
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
📄 www.nature.com/articles/s41...
💾 cluster.foldseek.com

📄 www.nature.com/articles/s41...
💾 cluster.foldseek.com
Our latest paper from the Baker lab is out today in mBio!
Interested in ocean hydrocarbon degradation & SIP-metagenomics?
Check this out!
👇
journals.asm.org/doi/10.1128/...
@archaeal.bsky.social

Our latest paper from the Baker lab is out today in mBio!
Interested in ocean hydrocarbon degradation & SIP-metagenomics?
Check this out!
👇
journals.asm.org/doi/10.1128/...
@archaeal.bsky.social



