@roman-bushuiev.bsky.social for receiving the Google PhD Fellowship 2025 in Health Research! 🎉💰 goo.gle/43wJWw8
@roman-bushuiev.bsky.social for receiving the Google PhD Fellowship 2025 in Health Research! 🎉💰 goo.gle/43wJWw8
Odpověď na to hledá projekt #PROTOCELL Kláry Hlouchové a @pluskal-lab.org, který vzniká v @iocbprague.bsky.social. 1/3
#ČeskáCestaDoVesmíru #CzechSpace #CzechSpaceWeek2025 #CzechSpaceWeek #UOCHB #IOCB #IOCBPrague

Odpověď na to hledá projekt #PROTOCELL Kláry Hlouchové a @pluskal-lab.org, který vzniká v @iocbprague.bsky.social. 1/3
#ČeskáCestaDoVesmíru #CzechSpace #CzechSpaceWeek2025 #CzechSpaceWeek #UOCHB #IOCB #IOCBPrague
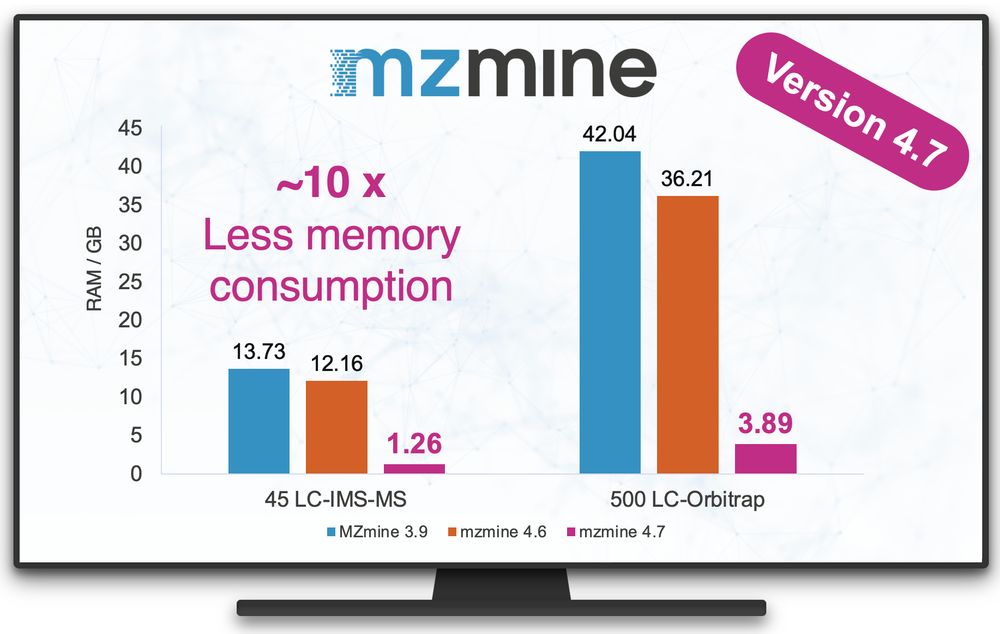
@robinschmid.bsky.social @pluskal-lab.org
www.nature.com/articles/s41...
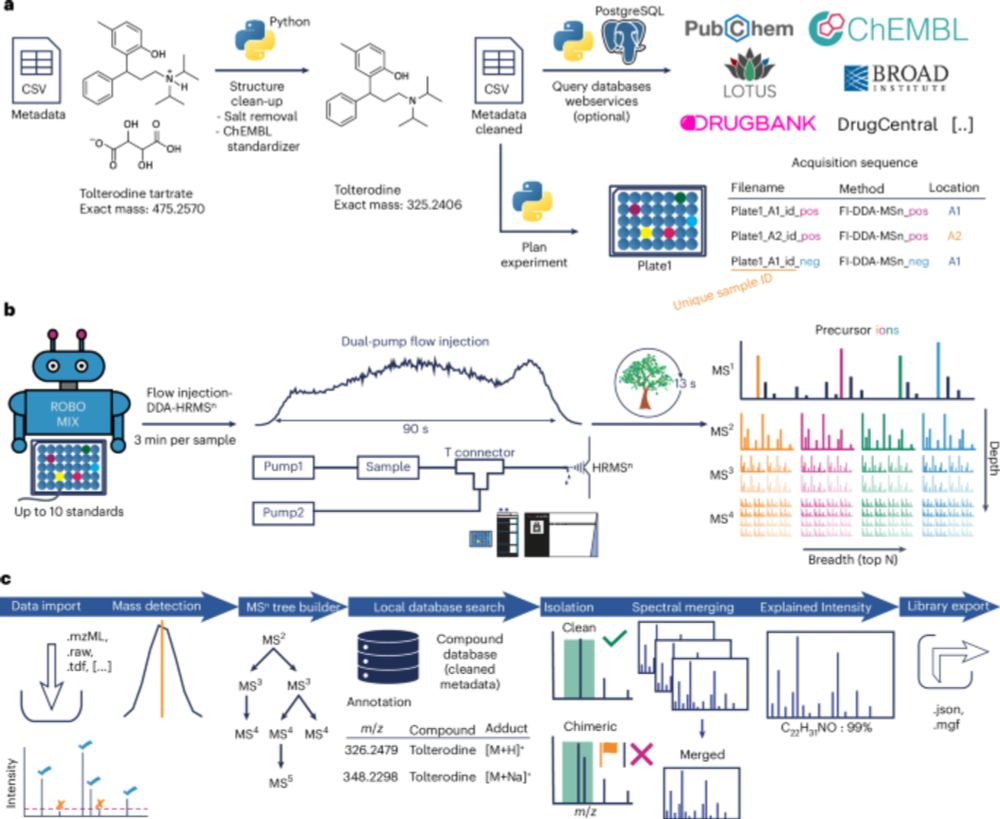
@robinschmid.bsky.social @pluskal-lab.org
www.nature.com/articles/s41...
A #research article about their vast #massspec library has been published in @natmethods.nature.com with Corinna Brungs & @robinschmidt.bsky.social as the first two authors.
See more ► www.uochb.cz/en/news/732





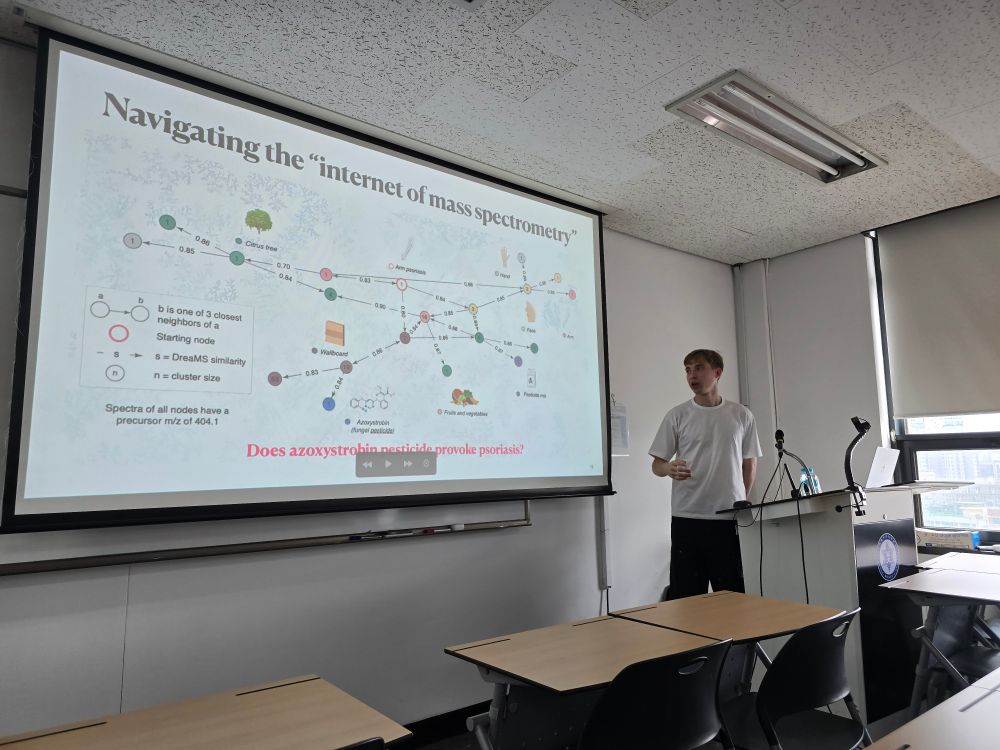

Compared to conventional LC-MS2, the data delivered:
- 84% more detected features
- 5.7× more MS² spectra
- 3× more spectral matches
Read the full poster to learn MoRE
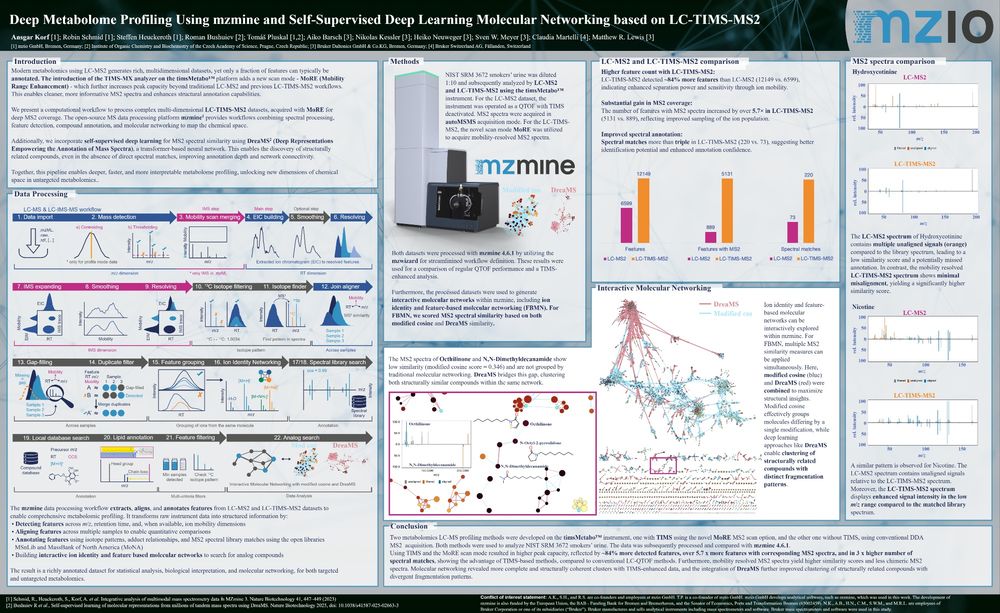
Compared to conventional LC-MS2, the data delivered:
- 84% more detected features
- 5.7× more MS² spectra
- 3× more spectral matches
Read the full poster to learn MoRE
🗓️ Submissions now open until June 13!
Two categoried:
👤 Human-Created // 🤖 AI-Generated
🏆Winner @ #MetSoc25 on June 24
Details👉 tinyurl.com/dh37xt78
📩info.emn@metabolomicssociety.org
Turn your #metabolomics research into visual art!
The categories:
👤 Human-made //🤖 AI-generated
🗓️ Submit Apr 1–May 31
🏆 Winners @ #MetSoc25 on June 24
Details 👉 tinyurl.com/dh37xt78
📩 info.emn@metabolomicssociety.org
#MetaboART #SciArt
@metsoc.bsky.social

🗓️ Submissions now open until June 13!
Two categoried:
👤 Human-Created // 🤖 AI-Generated
🏆Winner @ #MetSoc25 on June 24
Details👉 tinyurl.com/dh37xt78
📩info.emn@metabolomicssociety.org


www.nature.com/articles/s41...
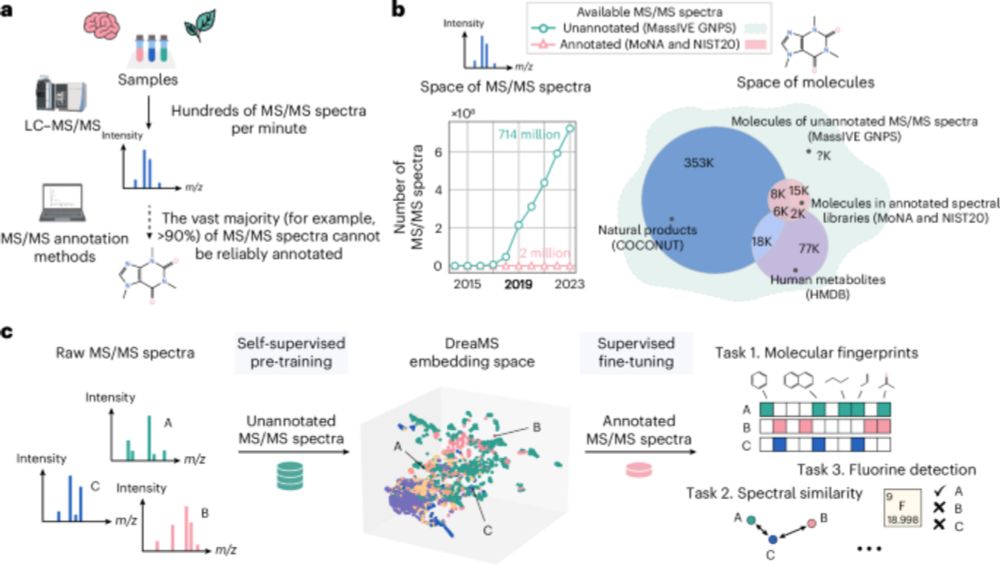
www.nature.com/articles/s41...

www.nature.com/articles/s41...

www.nature.com/articles/s41...
Led by experts @robinschmid.bsky.social and @ansgarkorf.bsky.social
𝗡𝗼 𝘀𝗶𝗴𝗻–𝘂𝗽 𝗿𝗲𝗾𝘂𝗶𝗿𝗲𝗱, 𝗷𝘂𝘀𝘁 𝗱𝗿𝗼𝗽 𝗯𝘆 𝘁𝗵𝗲 𝗯𝗼𝗼𝘁𝗵 𝗮𝘁 𝟭𝟮 𝗺𝗶𝗱𝗱𝗮𝘆.
See you there! 👋

Led by experts @robinschmid.bsky.social and @ansgarkorf.bsky.social
𝗡𝗼 𝘀𝗶𝗴𝗻–𝘂𝗽 𝗿𝗲𝗾𝘂𝗶𝗿𝗲𝗱, 𝗷𝘂𝘀𝘁 𝗱𝗿𝗼𝗽 𝗯𝘆 𝘁𝗵𝗲 𝗯𝗼𝗼𝘁𝗵 𝗮𝘁 𝟭𝟮 𝗺𝗶𝗱𝗱𝗮𝘆.
See you there! 👋
Our team will be showcasing our latest developments in #mzmine at this year’s conference.
Stop by Booth 210 for live demonstrations and insights into how you can streamline your data processing across multiple vendors, adding speed and confidence.

See more ► www.uochb.cz/en/open-posi... ⤵️
#openposition #iocbprague #iocb #researchjobs #biosciences

See more ► www.uochb.cz/en/open-posi... ⤵️
#openposition #iocbprague #iocb #researchjobs #biosciences
@iocbprague.bsky.social! Both new and advanced users are learning about the latest mzmine features from our amazing instructors @ansgarkorf.bsky.social, Josh Smith,
@titodamiani.bsky.social, and @roman-bushuiev.bsky.social.

@iocbprague.bsky.social! Both new and advanced users are learning about the latest mzmine features from our amazing instructors @ansgarkorf.bsky.social, Josh Smith,
@titodamiani.bsky.social, and @roman-bushuiev.bsky.social.

Rapid MALDI-MS/MS-Based Profiling of Lipid A Species from Gram-Negative Bacteria Utilizing Trapped Ion Mobility Spectrometry and mzmine (Rudt, Froning, @robinschmid.bsky.social, @pluskal-lab.org)–AC: doi.org/10.1021/acs....
@iocbprague.bsky.social @mzmine.bsky.social @uni-muenster.de
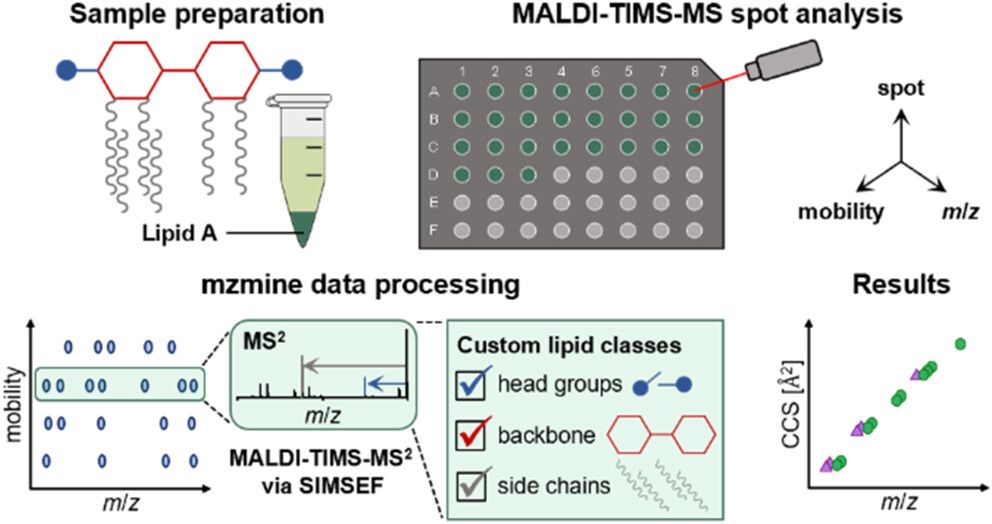
Rapid MALDI-MS/MS-Based Profiling of Lipid A Species from Gram-Negative Bacteria Utilizing Trapped Ion Mobility Spectrometry and mzmine (Rudt, Froning, @robinschmid.bsky.social, @pluskal-lab.org)–AC: doi.org/10.1021/acs....
@iocbprague.bsky.social @mzmine.bsky.social @uni-muenster.de
Presenters: Zdeněk Kameník (Laboratory of Antibiotic Resistance and Microbial Metabolomics IMIC);
Tito Damiani and Joshua Smith (Tomáš Pluskal group @iocbprague.bsky.social)
Learning new things #intermicro


Presenters: Zdeněk Kameník (Laboratory of Antibiotic Resistance and Microbial Metabolomics IMIC);
Tito Damiani and Joshua Smith (Tomáš Pluskal group @iocbprague.bsky.social)
Learning new things #intermicro
Registration:
docs.google.com/forms/d/e/1F...
Program:
docs.google.com/document/d/1...

Registration:
docs.google.com/forms/d/e/1F...
Program:
docs.google.com/document/d/1...
Computational metabolomics reveals overlooked chemodiversity of alkaloid scaffolds in Piper fimbriulatum
📖 doi.org/10.1111/tpj.70086
📝 Damiani, Smith et al @tomas_pluskal´s lab
🗺️ @iocbprague.bsky.social @czechacademy.bsky.social @CharlesUniPRG