🎓UT Austin 2020
Eliminate your opponents of course.
Recently, my friend @fernpizza.bsky.social showed how plasmids compete intracellularly (check out his paper published in Science today!). With @baym.lol, we now know they can fight.
www.biorxiv.org/content/10.1...


We obtained lots of thermal stable plastic degrading enzymes from the deep sea (Guaymas Basin, Gulf of California)

We obtained lots of thermal stable plastic degrading enzymes from the deep sea (Guaymas Basin, Gulf of California)


Free access link: rdcu.be/eLtCH
🧵 by @yifanzhou.bsky.social 👇
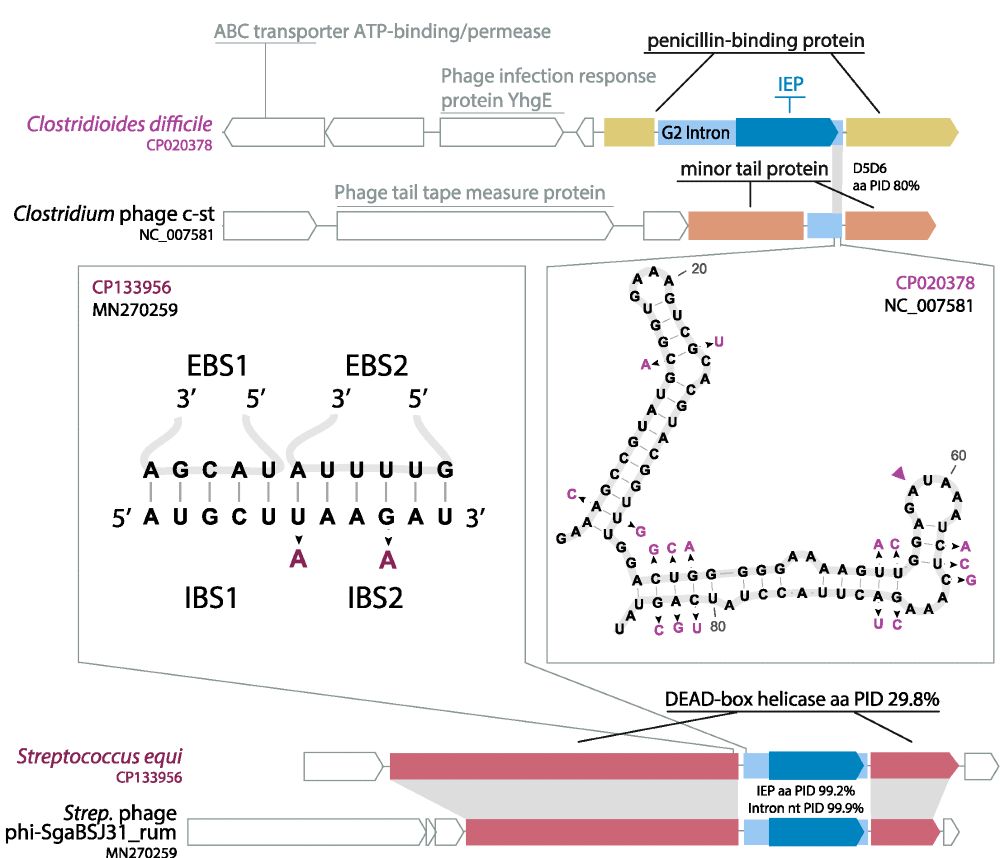
@mkrupovic.bsky.social @deemteam.bsky.social @anagtz.bsky.social
www.nature.com/articles/s41...

Free access link: rdcu.be/eLtCH
🧵 by @yifanzhou.bsky.social 👇
Read More: www.npr.org/2025/10/13/g...


Read More: www.npr.org/2025/10/13/g...

@shellyscrib.bsky.social @vscooper.micropopbio.org www.biorxiv.org/content/10.1...


asm.org/press-releas...

asm.org/press-releas...
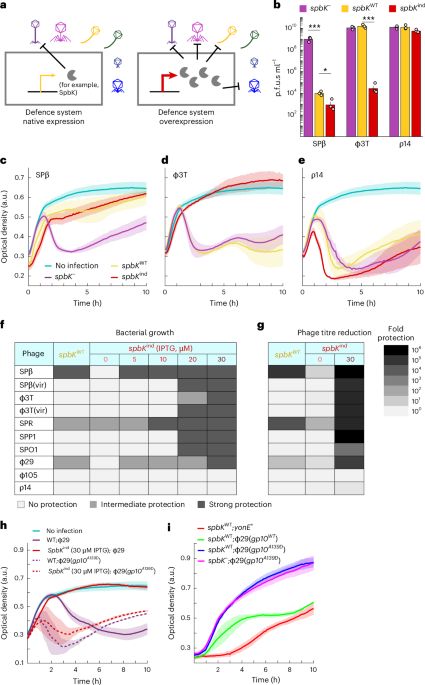
Phil. Trans. R. Soc. B by @seanmeaden.bsky.social, Edze Westra and @peterfineran.bsky.social
royalsocietypublishing.org/doi/10.1098/...

Phil. Trans. R. Soc. B by @seanmeaden.bsky.social, Edze Westra and @peterfineran.bsky.social
royalsocietypublishing.org/doi/10.1098/...
Good hands aren't some magic gift from the PCR gods, you have to develop them through directed repetitive practice, like any other skill
Good hands aren't some magic gift from the PCR gods, you have to develop them through directed repetitive practice, like any other skill