Structure biology| Drug resistance| Transporter| Cryo-EM| Membrane proteins|Microbiology|
Past: #UCSF, Research Specialist II | Bio5 postdoctoral fellow @uaarizona | #ICGEB #JNU #MSU

www.nature.com/articles/s41...

www.nature.com/articles/s41...
@ohsunews.bsky.social #OHSU @uofcalifornia.bsky.social #UCSF
@ohsunews.bsky.social #OHSU @uofcalifornia.bsky.social #UCSF
🙌 🎉 🥳
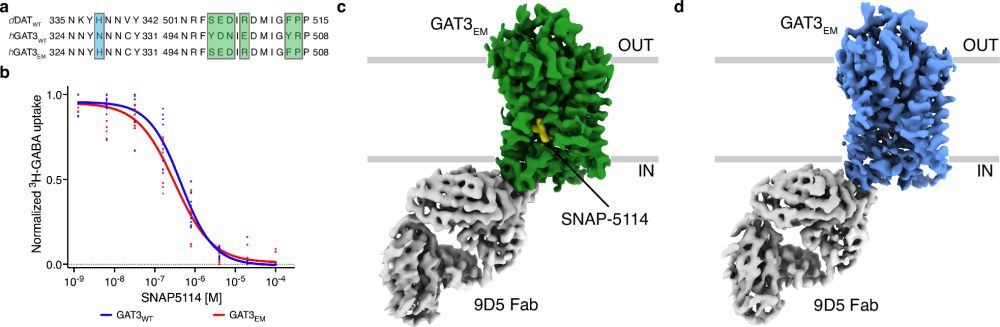
@nkhandelwal.bsky.social, Devin J Seka, Sree G Balasubramani, Miles S Dickinson, Alexander Myasnikov, @igech.bsky.social, Robert M Stroud.
🙌 🎉 🥳
@nkhandelwal.bsky.social, Devin J Seka, Sree G Balasubramani, Miles S Dickinson, Alexander Myasnikov, @igech.bsky.social, Robert M Stroud.
We’ve updated our preprint! It now covers the full MemBrain v2 pipeline for end-to-end membrane analysis in #CryoET: segmentation, particle picking, and spatial statistics.
🔗 Preprint: doi.org/10.1101/2024...
🔗 Code: github.com/CellArchLab/...
🧵(1/6) #TeamTomo
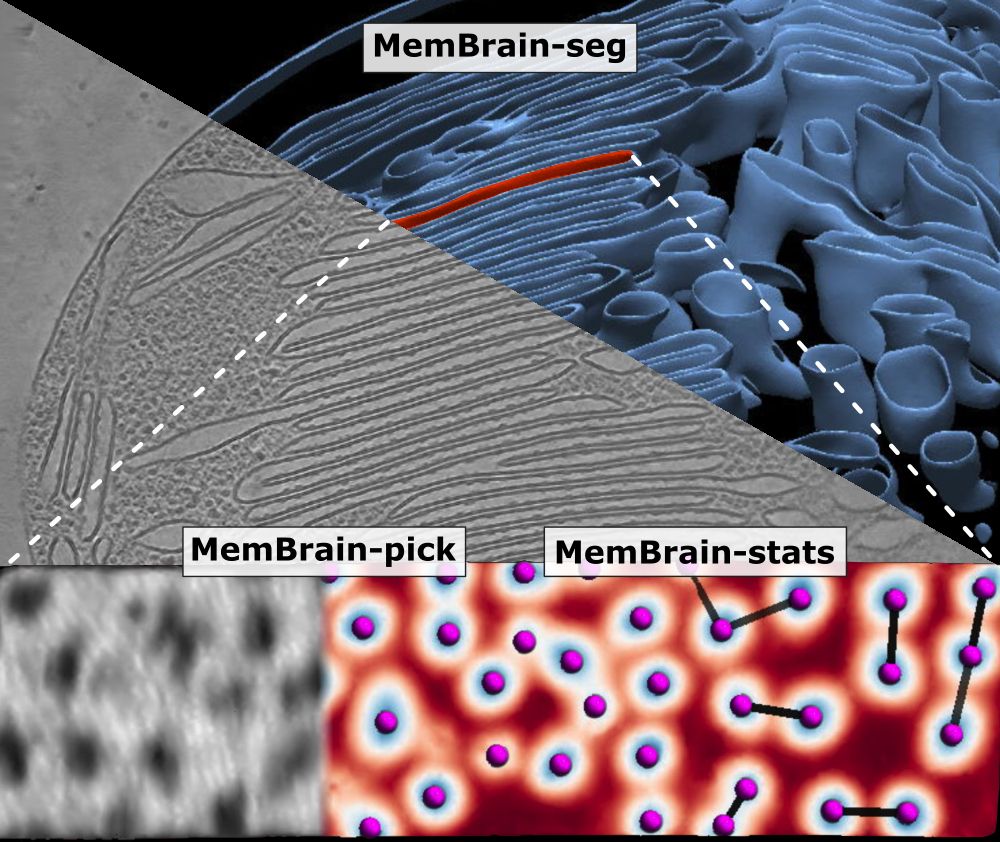
We’ve updated our preprint! It now covers the full MemBrain v2 pipeline for end-to-end membrane analysis in #CryoET: segmentation, particle picking, and spatial statistics.
🔗 Preprint: doi.org/10.1101/2024...
🔗 Code: github.com/CellArchLab/...
🧵(1/6) #TeamTomo
www.sciencedirect.com/science/arti...
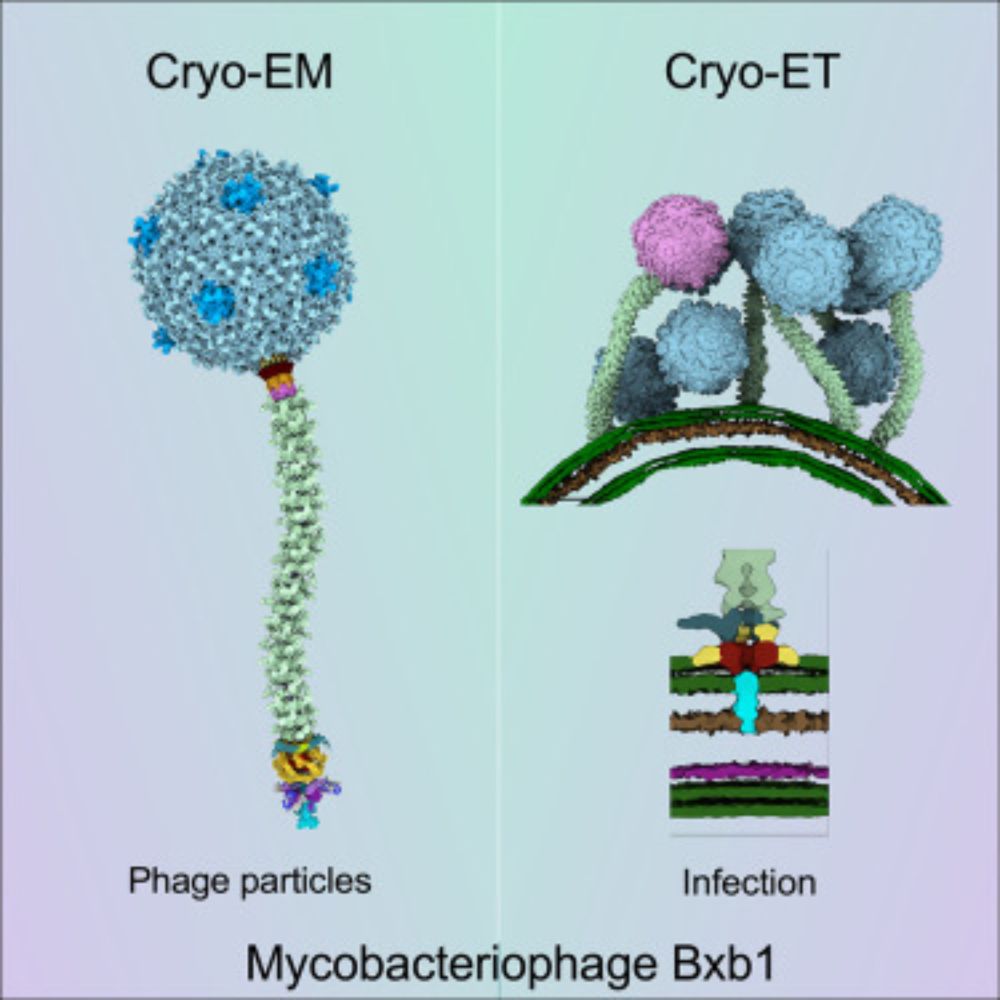
www.sciencedirect.com/science/arti...
Featuring a new automatic Micrograph Junk Detector that labels and rejects contaminants, new tools for curating subsets of particles, performance and stability fixes, and more!
Full changelog: cryosparc.com/updates
#cryoEM
Featuring a new automatic Micrograph Junk Detector that labels and rejects contaminants, new tools for curating subsets of particles, performance and stability fixes, and more!
Full changelog: cryosparc.com/updates
#cryoEM
www.nature.com/articles/s41...
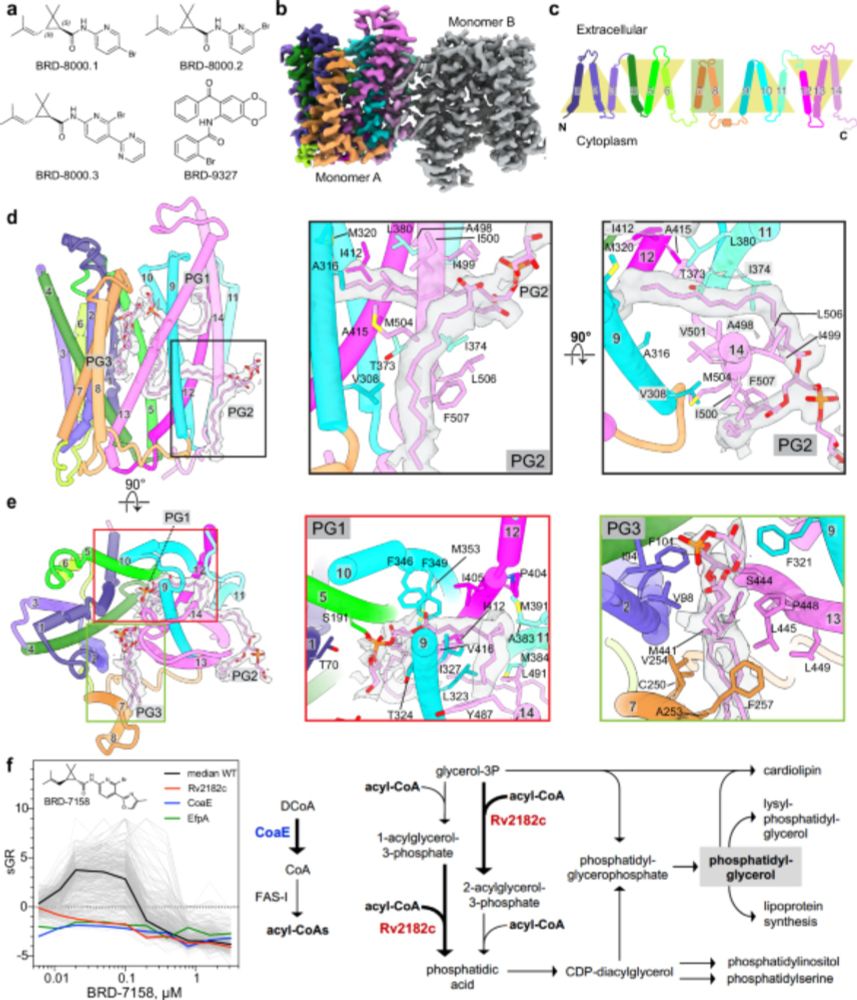
www.nature.com/articles/s41...
Coming to homebrew and flatpak soon!
- Clark & Labute comes to Coot 1, Moorhen & chapi!
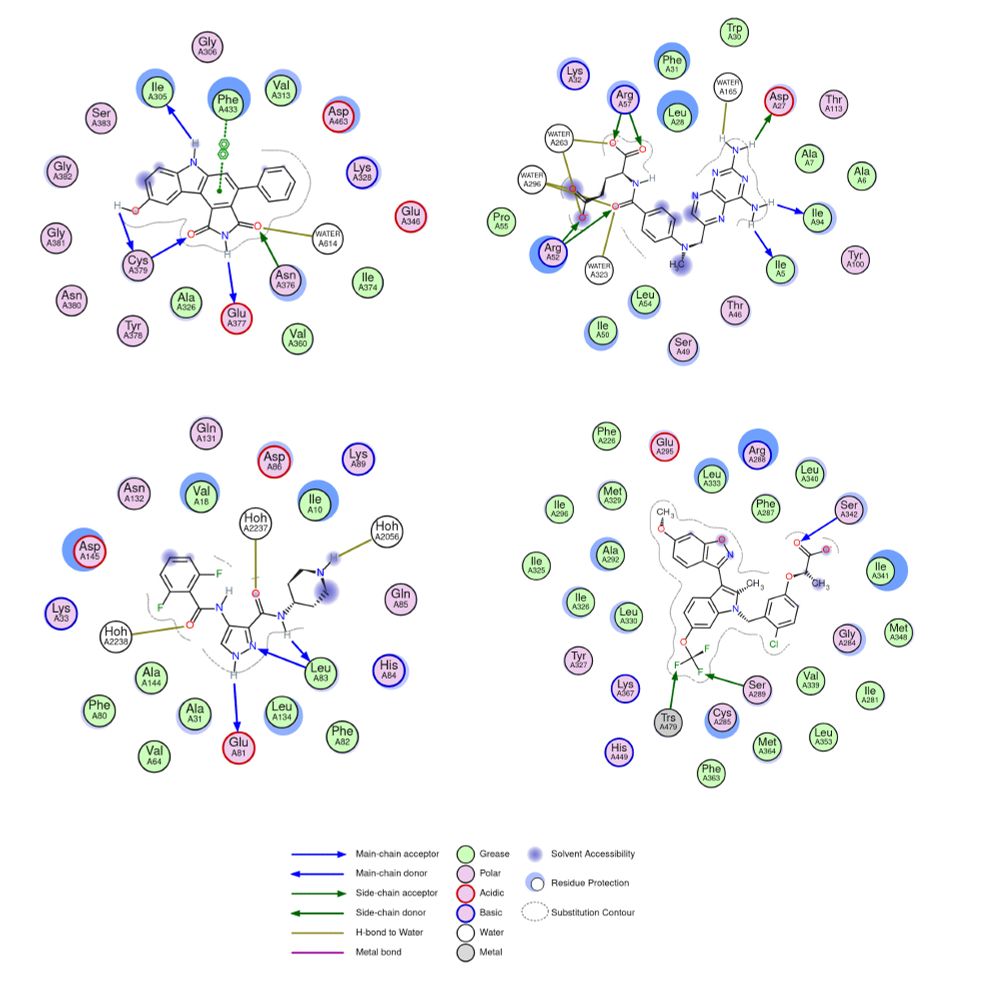
(svg, 2D ligand environment view)
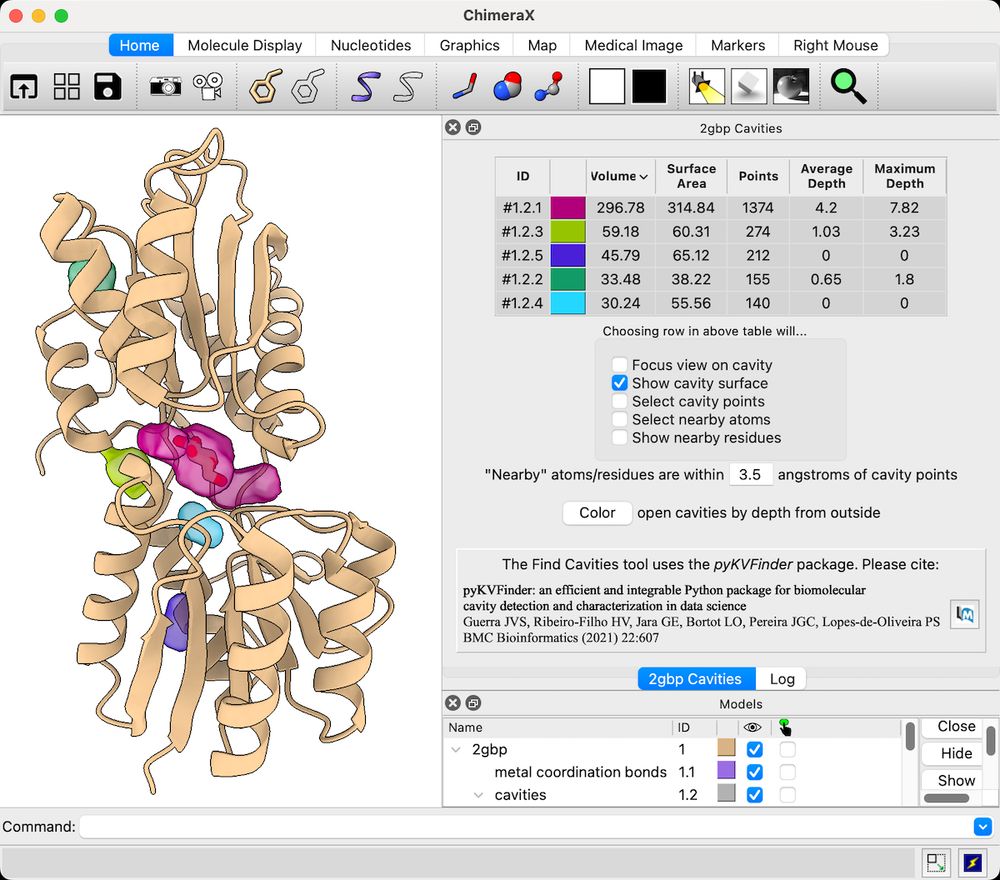
- Solid Symmetry (symmetry that you can see!)
- PAE plot for AlphaFold models
#coot
#molmatinf
Coming to homebrew and flatpak soon!
- Clark & Labute comes to Coot 1, Moorhen & chapi!
(svg, 2D ligand environment view)
- Solid Symmetry (symmetry that you can see!)
- PAE plot for AlphaFold models
#coot
#molmatinf
How do cells avoid the "kiss of death"?
Gap junctions share life-saving signals—but under stress, they shut down to prevent spreading cytotoxic damage.
#CryoEM reveals the molecular switches behind this process!
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
How do cells avoid the "kiss of death"?
Gap junctions share life-saving signals—but under stress, they shut down to prevent spreading cytotoxic damage.
#CryoEM reveals the molecular switches behind this process!
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
tinyurl.com/MGuptaSA
Huge congratulations to the team- @nkhandelwal.bsky.social, Andrew Nelson, Peter Hwang, Sergei Pourmal, Jeffrey Bennett, ROBERT M STROUD!

tinyurl.com/MGuptaSA
Huge congratulations to the team- @nkhandelwal.bsky.social, Andrew Nelson, Peter Hwang, Sergei Pourmal, Jeffrey Bennett, ROBERT M STROUD!

