Lab website: https://shorturl.at/lowCL
www.nature.com/articles/s43...

www.nature.com/articles/s43...
But we think it's because the metabolic consequences of mtDNA mutation are powerful drivers of tumor initiation and progression.
www.cell.com/trends/cance...

But we think it's because the metabolic consequences of mtDNA mutation are powerful drivers of tumor initiation and progression.
www.cell.com/trends/cance...
These mutations have never been seen in the human germline or in mito disease.
Clearly they are not compatible with life, but tumors are not only unfazed by this, they are happily selecting for the most deleterious mtDNA mutations observed on planet earth.
Why is that?
These mutations have never been seen in the human germline or in mito disease.
Clearly they are not compatible with life, but tumors are not only unfazed by this, they are happily selecting for the most deleterious mtDNA mutations observed on planet earth.
Why is that?
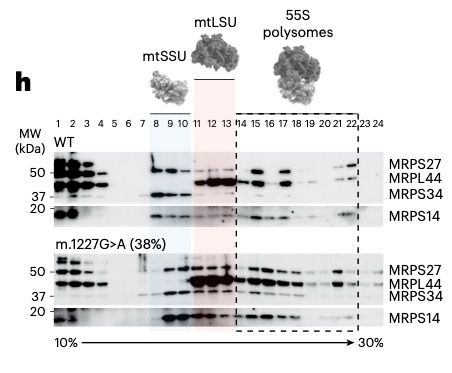
Most mtDNA mutations in cancer have never been studied, and many are 5-50% heteroplasmy (VAF).
Because of the low heteroplasmy, they are often discounted as non-functional passengers.
The discoveries described in this paper establish that this is not a safe assumption.
Most mtDNA mutations in cancer have never been studied, and many are 5-50% heteroplasmy (VAF).
Because of the low heteroplasmy, they are often discounted as non-functional passengers.
The discoveries described in this paper establish that this is not a safe assumption.