Lab website: https://shorturl.at/lowCL



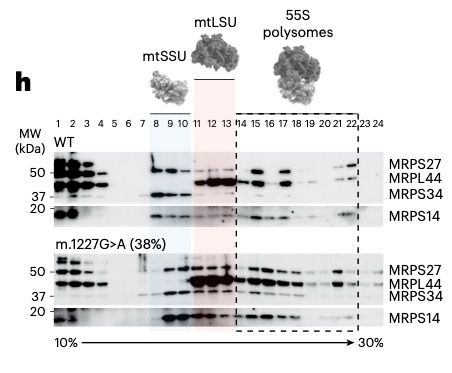







www.nature.com/articles/s42...

www.nature.com/articles/s42...



