
Account by lab members
- Menopause timing
- Age at Menarche
- Pelvic organ prolapse
The uterus isn’t just a passive player—it might hold the keys to predicting and preventing post-menopausal disorders.
7/8🧵

- Menopause timing
- Age at Menarche
- Pelvic organ prolapse
The uterus isn’t just a passive player—it might hold the keys to predicting and preventing post-menopausal disorders.
7/8🧵
But paired with histology, it can be.
We uncovered tissue-specific aging pathways:
🔻 Myometrium: ↓ ECM & muscle genes
🔺 Vaginal epithelium: ↓ epithelial +↑ immune genes
🔻 Ovarian cortex: ↓ angiogenesis
Organs don’t age as one.
6/8 🧵

But paired with histology, it can be.
We uncovered tissue-specific aging pathways:
🔻 Myometrium: ↓ ECM & muscle genes
🔺 Vaginal epithelium: ↓ epithelial +↑ immune genes
🔻 Ovarian cortex: ↓ angiogenesis
Organs don’t age as one.
6/8 🧵
This is more than known thinning: we saw extensive histological remodeling and sharp trajectory shifts at ~51 yrs.
❗Histology beats gene expression at capturing this transition.
5/8🧵

This is more than known thinning: we saw extensive histological remodeling and sharp trajectory shifts at ~51 yrs.
❗Histology beats gene expression at capturing this transition.
5/8🧵
Segmenting tissues with a vision transformer revealed that the myometrium (uterine muscle) is the most age-sensitive tissue.
Its histology transforms —collagen builds up, structure changes— with aging.
4/8🧵
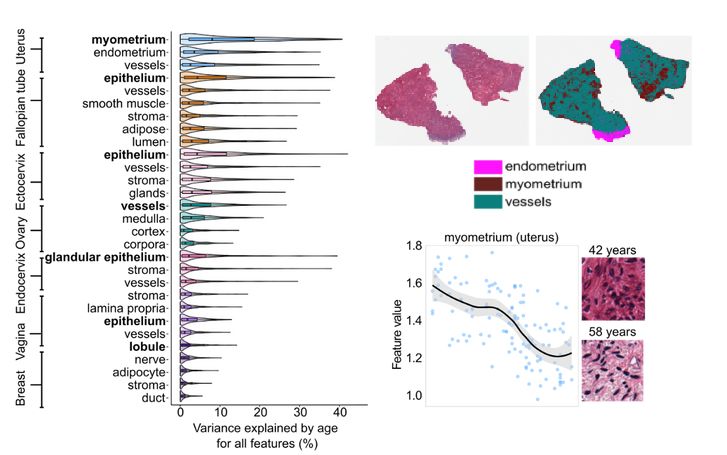
Segmenting tissues with a vision transformer revealed that the myometrium (uterine muscle) is the most age-sensitive tissue.
Its histology transforms —collagen builds up, structure changes— with aging.
4/8🧵
📈 Ovaries & vagina show gradual aging
⚡️ Uterus shows abrupt transition around age 51—right at menopause!
Transcriptomic data mirrored this: gene expression shifts sharply in the uterus but not in the ovary or vagina.
3/8🧵

📈 Ovaries & vagina show gradual aging
⚡️ Uterus shows abrupt transition around age 51—right at menopause!
Transcriptomic data mirrored this: gene expression shifts sharply in the uterus but not in the ovary or vagina.
3/8🧵
🖼️ 1,112 histological images
🧬 659 RNA-seq samples
🧠 Deep learning + vision transformers
📊 Multi-omics integration
All from 304 women aged 20-70 (GTEx dataset).
Across 7 organs: uterus, ovary, vagina, breast, ectocervix, fallopian tubes, and endocervix.
2/8🧵

🖼️ 1,112 histological images
🧬 659 RNA-seq samples
🧠 Deep learning + vision transformers
📊 Multi-omics integration
All from 304 women aged 20-70 (GTEx dataset).
Across 7 organs: uterus, ovary, vagina, breast, ectocervix, fallopian tubes, and endocervix.
2/8🧵
1️⃣ enable the discovery of novel transcripts that can not be found with GRCh38
2️⃣ contain regions not found in GRCh38 that harbor potentially novel genes (albeit at very low densities compared to the rest of the genome)
12

1️⃣ enable the discovery of novel transcripts that can not be found with GRCh38
2️⃣ contain regions not found in GRCh38 that harbor potentially novel genes (albeit at very low densities compared to the rest of the genome)
12
🌍More importantly, this increase is uneven between populations, with African individuals benefiting most (LWK, YRI).
10

🌍More importantly, this increase is uneven between populations, with African individuals benefiting most (LWK, YRI).
10
🔴We show that a population-diverse gene annotation increases the discovery of associations between SNPs and transcript expression, especially in non-European populations 🌍
7

🔴We show that a population-diverse gene annotation increases the discovery of associations between SNPs and transcript expression, especially in non-European populations 🌍
7
3

3

